Johnson_DOX_24_1
Omar Johnson
2024-07-17
Last updated: 2024-07-23
Checks: 7 0
Knit directory: DOX_24_Github/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240723) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 88c6686. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/Johnson_DOX_24_1.html
Ignored: analysis/Johnson_DOX_24_2.html
Ignored: analysis/Johnson_DOX_24_3.html
Ignored: analysis/Johnson_DOX_24_4.html
Ignored: analysis/Johnson_DOX_24_5.html
Ignored: analysis/Johnson_DOX_24_6.html
Ignored: analysis/Johnson_DOX_24_7.html
Ignored: analysis/Johnson_DOX_24_8.html
Ignored: analysis/Johnson_DOX_24_RUV_Limma.html
Untracked files:
Untracked: Fig_2.Rmd
Untracked: Fig_2.html
Untracked: analysis/Johnson_DOX_24_RUV_Limma.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Johnson_DOX_24_1.Rmd) and
HTML (docs/Johnson_DOX_24_1.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 88c6686 | Omar-Johnson | 2024-07-23 | Publish the initial files for myproject |
Load Libraries
Load data
Fig-1-D- Correlation heatmap
# correlation matrix
Prot_cor_matrix <- cor(RUVg_Log2_quantnormalized_all10samples %>% as.matrix())
# Heatmap 1.
pheatmap(Prot_cor_matrix,
color = colorRampPalette(c("blue", "white", "red"))(200),
display_numbers = TRUE)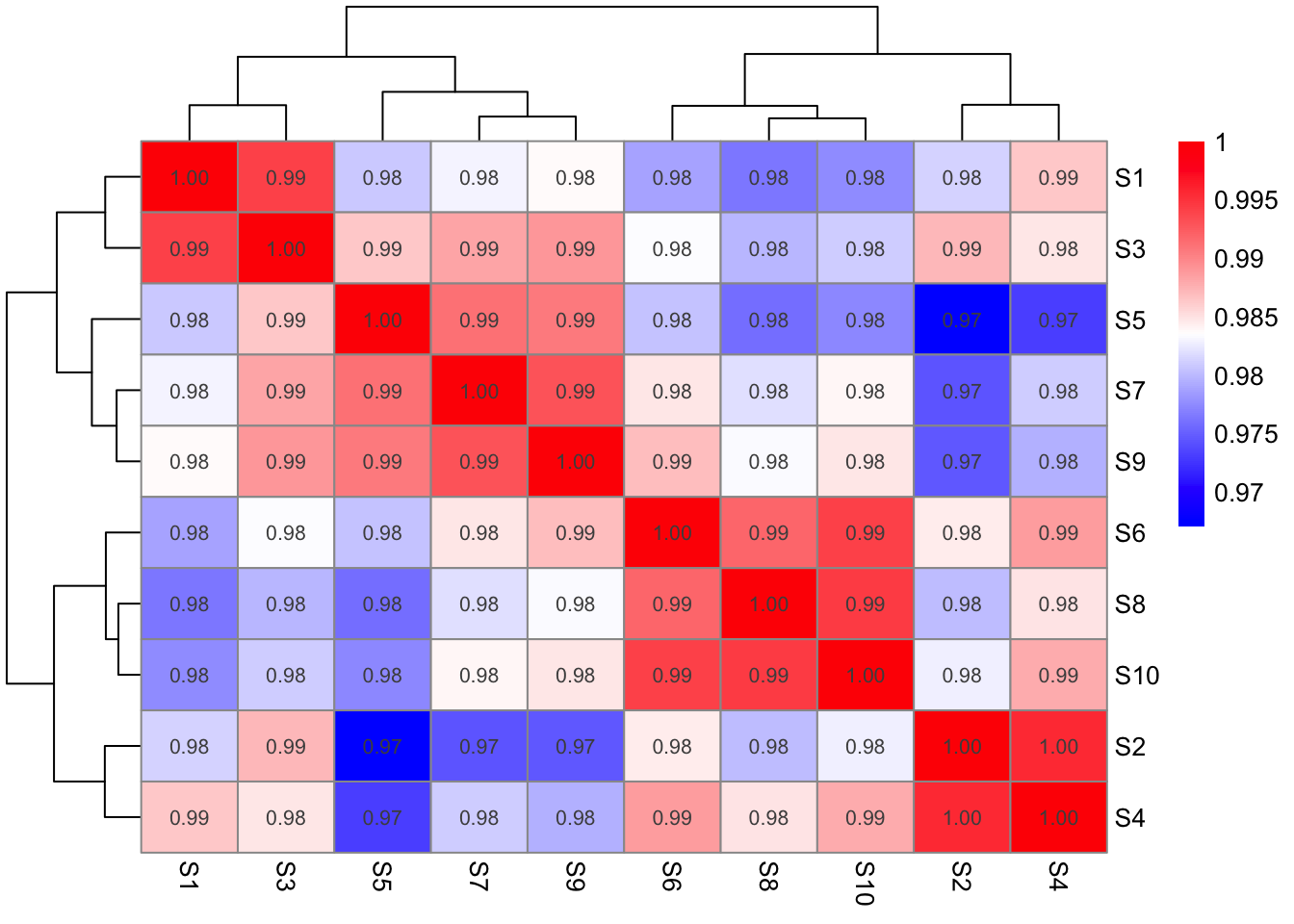
# Adjust column names
rownames(Prot_cor_matrix) <- Meta$Cond_Ind
colnames(Prot_cor_matrix) <- Meta$Cond_Ind
# Heatmap 2.
pheatmap(mat = Prot_cor_matrix,
color = colorRampPalette(c("blue", "white", "red"))(200),
display_numbers = TRUE
)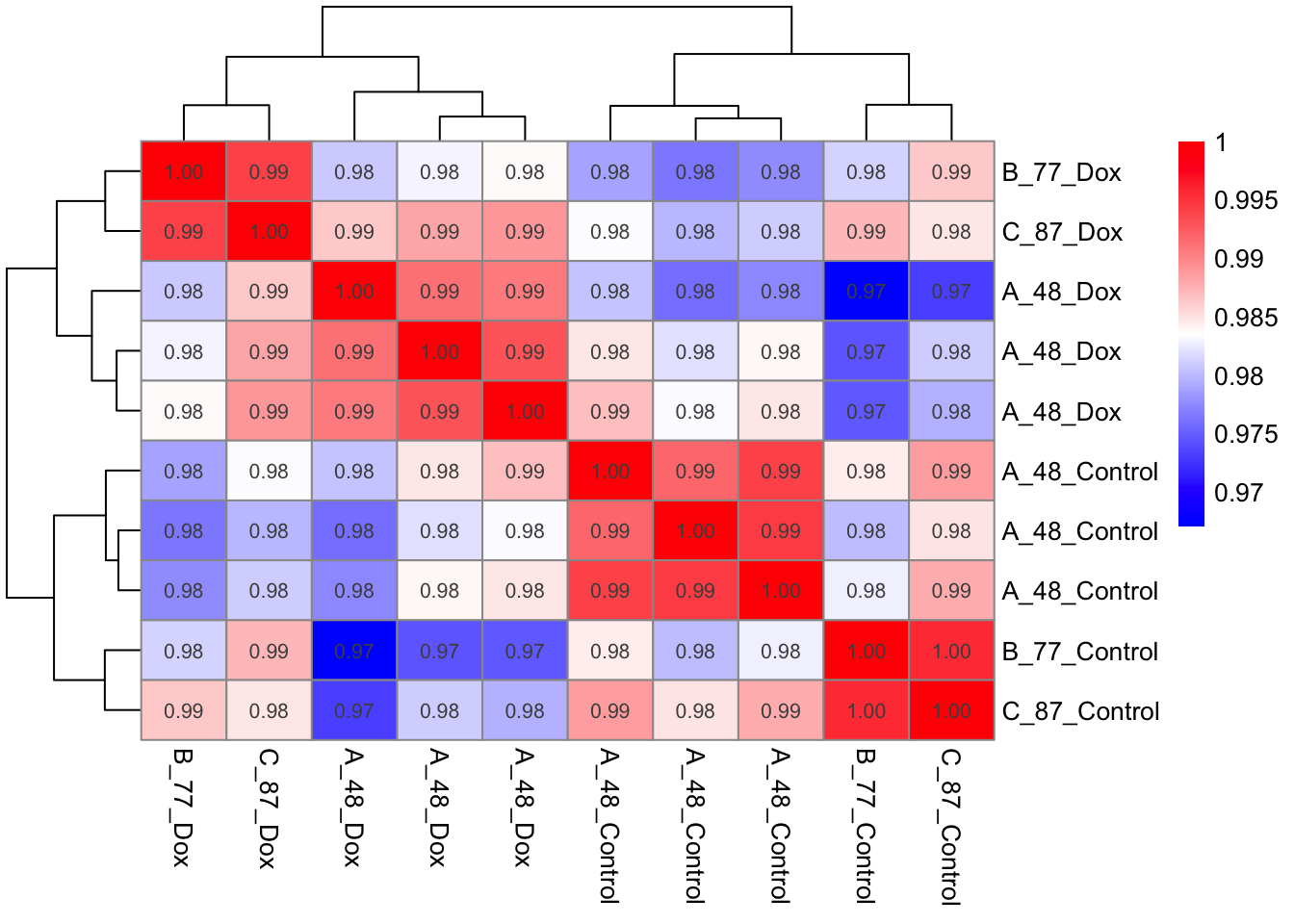
Fig-1-E iPSC-CM proteome compared to other tissue (GTEx)
Proteomics %>% head() Accession Entrez.Gene.ID Ensembl.Gene.ID Gene.Symbol
1 P53396 47 ENSG00000131473 ACLY
2 Q9H2P0 23394 ENSG00000101126 ADNP
3 Q96L96 57538 ENSG00000136383 ALPK3
4 Q15327 27063 ENSG00000148677 ANKRD1
5 Q92747 10552 ENSG00000241685 ARPC1A
6 O75787 10159 ENSG00000182220 ATP6AP2
Gene.ID
1 B4DIM0; B4E3P0; hsa:47; P53396; Q13037; Q9BRL0
2 E1P5Y2; hsa:23394; O94881; Q5BKU2; Q9H2P0; Q9UG34
3 hsa:57538; Q96L96; Q9P2L6
4 hsa:27063; Q15327; Q96LE7
5 A4D276; B4DLQ7; D6W5S1; hsa:10552; Q7Z5U8; Q86WU5; Q8IXQ0; Q92747
6 B7Z9I3; hsa:10159; O75787; Q5QTQ7; Q6T7F5; Q8NBP3; Q8NG15; Q96FV6; Q96LB5; Q9H2P8; Q9UG89
Abundance.Ratio...Veh.....Dox. Abundance.Ratio.Adj..P.Value...Veh.....Dox.
1 0.653 0.396029664
2 2.055 0.098818689
3 2.498 0.051655904
4 9.744 0.016363333
5 0.708 0.0960285
6 0.353 0.187490328
Abundances..Grouped...Dox Abundances..Grouped...Veh
1 19417357.35 12676982.35
2 3516801.084 7228452.172
3 1811200.791 4524233.176
4 6423990.119 62596866.19
5 53145253.16 37630913.11
6 8575431.426 3024303.718
Abundances..Grouped..CV......Dox Abundances..Grouped..CV......Veh
1 40.69 30.77
2 28.2 9.93
3 26.07 25.06
4 15.7 27.25
5 7.36 14
6 13.99 59.44
Abundance..F1..Sample..Dox..n.a Abundance..F3..Sample..Dox..n.a
1 14402176.63 9366275.609
2 4618733.906 3370283.281
3 863022.1563 1999058.094
4 3826071.625 6398937.516
5 48717851.41 41945664.09
6 6576761.375 7016359.25
Abundance..F5..Sample..Dox..Dox Abundance..F7..Sample..Dox..Dox
1 29436543.88 22451050.16
2 2997374.313 2408716.5
3 545111.9688 1405675.656
4 4398013.953 5165224.781
5 50559977.91 50742792.91
6 9019724.875 9206177
Abundance..F9..Sample..Dox..Dox Abundance..F2..Control..Veh..n.a
1 17300954.38 6913680.227
2 1941331.219 7074523.469
3 1021132.875 2214027.172
4 4080027.609 52421690.72
5 48816880.66 39477613.69
6 7912307.188 2709389.25
Abundance..F4..Control..Veh..n.a Abundance..F6..Control..Veh..Veh
1 5406406.719 10494270.92
2 5397867.813 6444366.813
3 5356827.313 4077320.656
4 38865174.44 62303686.18
5 36143313.94 27690129.17
6 5919575.719 2184336.5
Abundance..F8..Control..Veh..Veh Abundance..F10..Control..Veh..Veh
1 14124540.34 12160232.45
2 7831825.969 6710694.406
3 4393884.313 3877961.406
4 81896769 69853076.38
5 36418791.34 30344995.19
6 1238241.469 2856388.75Gtex %>% head() gene.id Adrenal.Gland Artery Brain.Cerebellum Breast Colon
1 ENSG00000000003 -0.71 -0.95 -2.28 -0.37 -0.46
2 ENSG00000000419 0.32 -0.60 -1.22 -0.05 -0.60
3 ENSG00000000457 -0.69 NA NA NA 0.33
4 ENSG00000000938 -1.23 -0.07 -1.40 -1.13 -1.49
5 ENSG00000000971 -1.03 2.09 -3.35 0.89 -1.46
6 ENSG00000001036 0.74 -1.14 -2.59 -0.41 0.42
GE.junction Esophagus.Muscle Heart.Atrial Heart.Ventricle Liver Lung
1 -1.17 -1.94 -1.45 -1.44 0.15 -0.30
2 -0.27 -0.54 -0.43 -0.19 0.49 -0.04
3 0.10 -0.40 0.19 0.48 -0.37 -0.53
4 -0.84 -0.97 -0.52 -1.39 -0.16 1.25
5 0.14 -0.14 0.61 -1.02 -0.57 0.02
6 -0.98 -1.23 -0.12 -0.29 0.31 -0.34
Minor.Salivary Muscle.Skeletal Nerve.Tibial Ovary Pancreas Pituitary Prostate
1 -1.35 -1.15 0.75 0.28 1.09 -0.45 -1.04
2 0.29 -0.52 -0.70 0.07 1.74 -0.35 -0.16
3 NA 0.71 -0.69 -1.07 -0.13 -0.63 0.90
4 -0.90 -2.39 -1.67 -3.17 -1.23 -2.27 -1.57
5 -1.54 -1.46 1.62 0.32 -2.73 -0.82 -1.56
6 -0.46 -1.65 -0.62 -0.48 -0.42 0.57 -0.60
Skin Small.Intestine Spleen Stomach Testis Thyroid Uterus Vagina
1 -0.45 -0.74 -1.74 0.20 -0.32 -1.00 -1.06 -1.25
2 -0.33 -0.49 -0.26 0.40 0.25 0.42 -0.18 -0.23
3 -0.65 0.18 -0.12 0.66 -1.09 -0.69 NA 0.21
4 -1.55 -0.52 2.20 -1.71 -2.30 -1.76 -1.99 -0.43
5 0.49 -2.21 -1.99 -1.31 -0.82 -1.71 -1.16 -0.50
6 -0.67 0.84 -0.08 0.20 -0.37 2.13 -0.58 -0.55Gtex_genelist %>% head() X entrezgene_id ensembl_gene_id hgnc_symbol
1 1 7105 ENSG00000000003 TSPAN6
2 2 8813 ENSG00000000419 DPM1
3 3 57147 ENSG00000000457 SCYL3
4 4 2268 ENSG00000000938 FGR
5 5 3075 ENSG00000000971 CFH
6 6 2519 ENSG00000001036 FUCA2Protein_list <- Proteomics %>%
dplyr::select(Entrez.Gene.ID,Abundance..F1..Sample..Dox..n.a, Abundance..F3..Sample..Dox..n.a,Abundance..F5..Sample..Dox..Dox,Abundance..F7..Sample..Dox..Dox,Abundance..F9..Sample..Dox..Dox,Abundance..F2..Control..Veh..n.a,Abundance..F4..Control..Veh..n.a,Abundance..F6..Control..Veh..Veh,Abundance..F8..Control..Veh..Veh,Abundance..F10..Control..Veh..Veh) %>%
mutate(Abundance..F1..Sample..Dox..n.a=as.numeric(Abundance..F1..Sample..Dox..n.a)) %>%
mutate(Abundance..F3..Sample..Dox..n.a=as.numeric(Abundance..F3..Sample..Dox..n.a)) %>%
mutate(Abundance..F5..Sample..Dox..Dox=as.numeric(Abundance..F5..Sample..Dox..Dox)) %>%
mutate(Abundance..F7..Sample..Dox..Dox=as.numeric(Abundance..F7..Sample..Dox..Dox)) %>%
mutate(Abundance..F9..Sample..Dox..Dox=as.numeric(Abundance..F9..Sample..Dox..Dox)) %>%
mutate(Abundance..F2..Control..Veh..n.a=as.numeric(Abundance..F2..Control..Veh..n.a)) %>%
mutate(Abundance..F4..Control..Veh..n.a=as.numeric(Abundance..F4..Control..Veh..n.a)) %>%
mutate(Abundance..F6..Control..Veh..Veh=as.numeric(Abundance..F6..Control..Veh..Veh)) %>%
mutate(Abundance..F8..Control..Veh..Veh=as.numeric(Abundance..F8..Control..Veh..Veh)) %>%
mutate(Abundance..F10..Control..Veh..Veh=as.numeric(Abundance..F10..Control..Veh..Veh)) %>%
mutate("log2_abundance_77-1_Dox"= log2(Abundance..F1..Sample..Dox..n.a)) %>%
mutate("log2_abundance_87-1_Dox"= log2(Abundance..F3..Sample..Dox..n.a)) %>%
mutate("log2_abundance_048-A_1_Dox"= log2(Abundance..F5..Sample..Dox..Dox)) %>%
mutate("log2_abundance_048-A_2_Dox"= log2(Abundance..F7..Sample..Dox..Dox)) %>%
mutate("log2_abundance_048-A_3_Dox"= log2(Abundance..F9..Sample..Dox..Dox)) %>%
mutate("log2_abundance_77-1_Veh"= log2(Abundance..F2..Control..Veh..n.a)) %>%
mutate("log2_abundance_87-1_Veh"= log2(Abundance..F4..Control..Veh..n.a)) %>%
mutate("log2_abundance_048-A_1_Veh"= log2(Abundance..F6..Control..Veh..Veh)) %>%
mutate("log2_abundance_048-A_2_Veh"= log2(Abundance..F8..Control..Veh..Veh)) %>%
mutate("log2_abundance_048-A_3_Veh"= log2(Abundance..F10..Control..Veh..Veh)) %>%
mutate(Entrez.Gene.ID=as.numeric(Entrez.Gene.ID)) %>%
na.omit(.)Warning: There was 1 warning in `mutate()`.
ℹ In argument: `Abundance..F1..Sample..Dox..n.a =
as.numeric(Abundance..F1..Sample..Dox..n.a)`.
Caused by warning:
! NAs introduced by coercionWarning: There was 1 warning in `mutate()`.
ℹ In argument: `Abundance..F3..Sample..Dox..n.a =
as.numeric(Abundance..F3..Sample..Dox..n.a)`.
Caused by warning:
! NAs introduced by coercionWarning: There was 1 warning in `mutate()`.
ℹ In argument: `Abundance..F5..Sample..Dox..Dox =
as.numeric(Abundance..F5..Sample..Dox..Dox)`.
Caused by warning:
! NAs introduced by coercionWarning: There was 1 warning in `mutate()`.
ℹ In argument: `Abundance..F7..Sample..Dox..Dox =
as.numeric(Abundance..F7..Sample..Dox..Dox)`.
Caused by warning:
! NAs introduced by coercionWarning: There was 1 warning in `mutate()`.
ℹ In argument: `Abundance..F9..Sample..Dox..Dox =
as.numeric(Abundance..F9..Sample..Dox..Dox)`.
Caused by warning:
! NAs introduced by coercionWarning: There was 1 warning in `mutate()`.
ℹ In argument: `Abundance..F2..Control..Veh..n.a =
as.numeric(Abundance..F2..Control..Veh..n.a)`.
Caused by warning:
! NAs introduced by coercionWarning: There was 1 warning in `mutate()`.
ℹ In argument: `Abundance..F4..Control..Veh..n.a =
as.numeric(Abundance..F4..Control..Veh..n.a)`.
Caused by warning:
! NAs introduced by coercionWarning: There was 1 warning in `mutate()`.
ℹ In argument: `Abundance..F6..Control..Veh..Veh =
as.numeric(Abundance..F6..Control..Veh..Veh)`.
Caused by warning:
! NAs introduced by coercionWarning: There was 1 warning in `mutate()`.
ℹ In argument: `Abundance..F8..Control..Veh..Veh =
as.numeric(Abundance..F8..Control..Veh..Veh)`.
Caused by warning:
! NAs introduced by coercionWarning: There was 1 warning in `mutate()`.
ℹ In argument: `Abundance..F10..Control..Veh..Veh =
as.numeric(Abundance..F10..Control..Veh..Veh)`.
Caused by warning:
! NAs introduced by coercionWarning: There was 1 warning in `mutate()`.
ℹ In argument: `Entrez.Gene.ID = as.numeric(Entrez.Gene.ID)`.
Caused by warning:
! NAs introduced by coercion# Gtex Dataframe ----------------------------------------------------------
Convert <- Gtex %>% full_join(., Gtex_genelist, by=c("gene.id"="ensembl_gene_id"))
Convert_List<- Protein_list %>% left_join (., Convert, by=c("Entrez.Gene.ID"="entrezgene_id"), relationship = "many-to-many") %>%
distinct(Entrez.Gene.ID,.keep_all = TRUE) %>%
column_to_rownames("Entrez.Gene.ID") %>%
dplyr::select(!c("Abundance..F1..Sample..Dox..n.a", "Abundance..F3..Sample..Dox..n.a","Abundance..F5..Sample..Dox..Dox","Abundance..F7..Sample..Dox..Dox","Abundance..F9..Sample..Dox..Dox","Abundance..F2..Control..Veh..n.a","Abundance..F4..Control..Veh..n.a","Abundance..F6..Control..Veh..Veh","Abundance..F8..Control..Veh..Veh","Abundance..F10..Control..Veh..Veh","hgnc_symbol", "gene.id"))
Test <- rcorr
Test<- rcorr(as.matrix(Convert_List), type = "spearman")
Test<- rcorr(as.matrix(Convert_List), type = "pearson")
pheatmap(Test$r, display_numbers = TRUE)-1.png)
pheatmap(Test$r, display_numbers = FALSE)-2.png)
Fig-1-F Median abundance of cardiac proteins
RUVg_Log2_quantnormalized_all10samples_unlogged <- RUVg_Log2_quantnormalized_all10samples^2
RUVg_Log2_quantnormalized_all10samples_unlogged %>% head() S1 S3 S5 S7 S9 S2 S4
A0A0B4J2A2 328.5554 326.9973 344.8820 349.3119 352.7209 333.5604 343.2972
A0A0B4J2D5 575.3541 580.4938 574.6771 568.2429 573.3663 571.9707 564.5982
A0A494C071 319.5122 317.2598 303.3583 322.1064 317.5238 328.8213 328.9263
A0AVT1 174.4011 178.2046 205.8086 196.3262 195.1595 173.2376 180.2296
A0FGR8 411.5794 406.2913 397.4114 401.4574 400.3695 405.4398 406.3051
A0JLT2 324.3169 318.3035 315.9126 304.1986 322.8292 324.8402 324.0475
S6 S8 S10
A0A0B4J2A2 343.7512 340.3927 346.0124
A0A0B4J2D5 564.3974 562.3821 559.3047
A0A494C071 316.6769 326.8686 312.6095
A0AVT1 195.4427 178.4695 182.9383
A0FGR8 403.6977 405.8293 402.7064
A0JLT2 309.3706 297.4752 323.7055Heartspecpro <- RUVg_Log2_quantnormalized_all10samples_unlogged[c("P12883", "Q8WZ42", "Q14896","P13533", "P35609", "P45379", "P10916", "P27797", "P19429", "Q92736", "Q14524"), ]
Heartspecpro_matrix <- Heartspecpro %>% as.matrix()
Median_abundances <- Heartspecpro_matrix %>% rowMedians()Warning: useNames = NA is deprecated. Instead, specify either useNames = TRUE
or useNames = TRUE.Heartspecpro$Median_abundances <- Median_abundances
Heartspecpro$names <- c("P12883", "Q8WZ42", "Q14896","P13533", "P35609", "P45379", "P10916", "P27797", "P19429", "Q92736", "Q14524")
Heartspecpro$genes <- c("MYH7", "TTN", "MYBPC3", "MYH6", "ACTN2", "TNNT2", "MYL2", "CALR", "TNNI3", "RYR2", "SCN5A")
Heartspecpro$genes <- factor(x = Heartspecpro$genes, levels = rev(c("ACTN2","CALR","MYBPC3", "MYH6","MYH7","MYL2","RYR2","SCN5A", "TNNI3", "TNNT2","TTN")))
ggplot(Heartspecpro, aes(x = 1, y = genes, fill = Median_abundances)) +
geom_tile(color = "black", size = 0.5) + # Tiles with borders
scale_fill_gradient(low = "white", high = "red", limits = c(300, 750)) + # Gradient fill
geom_text(aes(label = paste(genes, Median_abundances, sep = "\n")), color = "black", size = 3) +
theme_minimal() +
theme(
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1),
axis.text.y = element_blank(),
axis.ticks.y = element_blank())Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.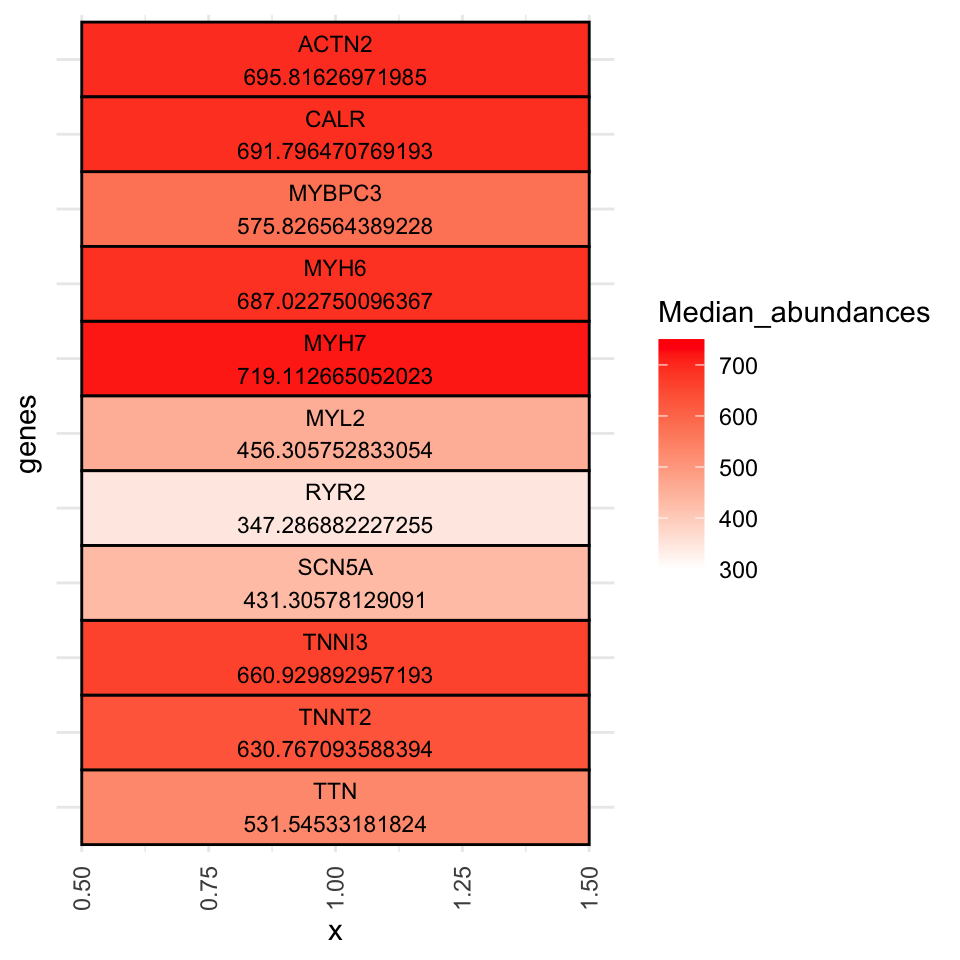
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur/Monterey 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] Hmisc_5.1-0
[2] ggraph_2.1.0
[3] igraph_1.5.1
[4] ReactomePA_1.40.0
[5] impute_1.70.0
[6] WGCNA_1.72-1
[7] fastcluster_1.2.3
[8] dynamicTreeCut_1.63-1
[9] BioNERO_1.4.2
[10] reshape2_1.4.4
[11] ggridges_0.5.4
[12] biomaRt_2.52.0
[13] ggvenn_0.1.10
[14] UpSetR_1.4.0
[15] DOSE_3.22.1
[16] variancePartition_1.26.0
[17] clusterProfiler_4.4.4
[18] pheatmap_1.0.12
[19] qvalue_2.28.0
[20] Homo.sapiens_1.3.1
[21] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[22] org.Hs.eg.db_3.15.0
[23] GO.db_3.15.0
[24] OrganismDbi_1.38.1
[25] GenomicFeatures_1.48.4
[26] AnnotationDbi_1.58.0
[27] cluster_2.1.4
[28] ggfortify_0.4.16
[29] lubridate_1.9.2
[30] forcats_1.0.0
[31] stringr_1.5.0
[32] dplyr_1.1.2
[33] purrr_1.0.2
[34] readr_2.1.4
[35] tidyr_1.3.0
[36] tibble_3.2.1
[37] ggplot2_3.4.3
[38] tidyverse_2.0.0
[39] RColorBrewer_1.1-3
[40] RUVSeq_1.30.0
[41] edgeR_3.38.4
[42] limma_3.52.4
[43] EDASeq_2.30.0
[44] ShortRead_1.54.0
[45] GenomicAlignments_1.32.1
[46] SummarizedExperiment_1.26.1
[47] MatrixGenerics_1.8.1
[48] matrixStats_1.0.0
[49] Rsamtools_2.12.0
[50] GenomicRanges_1.48.0
[51] Biostrings_2.64.1
[52] GenomeInfoDb_1.32.4
[53] XVector_0.36.0
[54] IRanges_2.30.1
[55] S4Vectors_0.34.0
[56] BiocParallel_1.30.4
[57] Biobase_2.56.0
[58] BiocGenerics_0.42.0
[59] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] rappdirs_0.3.3 rtracklayer_1.56.1 minet_3.54.0
[4] R.methodsS3_1.8.2 coda_0.19-4 bit64_4.0.5
[7] knitr_1.43 aroma.light_3.26.0 DelayedArray_0.22.0
[10] R.utils_2.12.2 rpart_4.1.19 data.table_1.14.8
[13] hwriter_1.3.2.1 KEGGREST_1.36.3 RCurl_1.98-1.12
[16] doParallel_1.0.17 generics_0.1.3 preprocessCore_1.58.0
[19] callr_3.7.3 RhpcBLASctl_0.23-42 RSQLite_2.3.1
[22] shadowtext_0.1.2 bit_4.0.5 tzdb_0.4.0
[25] enrichplot_1.16.2 xml2_1.3.5 httpuv_1.6.11
[28] viridis_0.6.4 xfun_0.40 hms_1.1.3
[31] jquerylib_0.1.4 evaluate_0.21 promises_1.2.1
[34] fansi_1.0.4 restfulr_0.0.15 progress_1.2.2
[37] caTools_1.18.2 dbplyr_2.3.3 htmlwidgets_1.6.2
[40] DBI_1.1.3 ggnewscale_0.4.9 backports_1.4.1
[43] annotate_1.74.0 aod_1.3.2 deldir_1.0-9
[46] vctrs_0.6.3 abind_1.4-5 cachem_1.0.8
[49] withr_2.5.0 ggforce_0.4.1 checkmate_2.2.0
[52] treeio_1.20.2 prettyunits_1.1.1 ape_5.7-1
[55] lazyeval_0.2.2 crayon_1.5.2 genefilter_1.78.0
[58] labeling_0.4.2 pkgconfig_2.0.3 tweenr_2.0.2
[61] nlme_3.1-163 nnet_7.3-19 rlang_1.1.1
[64] lifecycle_1.0.3 downloader_0.4 filelock_1.0.2
[67] BiocFileCache_2.4.0 rprojroot_2.0.3 polyclip_1.10-4
[70] graph_1.74.0 Matrix_1.5-4.1 aplot_0.2.0
[73] NetRep_1.2.7 boot_1.3-28.1 base64enc_0.1-3
[76] GlobalOptions_0.1.2 whisker_0.4.1 processx_3.8.2
[79] png_0.1-8 viridisLite_0.4.2 rjson_0.2.21
[82] bitops_1.0-7 getPass_0.2-2 R.oo_1.25.0
[85] ggnetwork_0.5.12 KernSmooth_2.23-22 blob_1.2.4
[88] shape_1.4.6 jpeg_0.1-10 gridGraphics_0.5-1
[91] reactome.db_1.81.0 scales_1.2.1 graphite_1.42.0
[94] memoise_2.0.1 magrittr_2.0.3 plyr_1.8.8
[97] gplots_3.1.3 zlibbioc_1.42.0 compiler_4.2.0
[100] scatterpie_0.2.1 BiocIO_1.6.0 clue_0.3-64
[103] intergraph_2.0-3 lme4_1.1-34 cli_3.6.1
[106] patchwork_1.1.3 ps_1.7.5 htmlTable_2.4.1
[109] Formula_1.2-5 mgcv_1.9-0 MASS_7.3-60
[112] tidyselect_1.2.0 stringi_1.7.12 highr_0.10
[115] yaml_2.3.7 GOSemSim_2.22.0 locfit_1.5-9.8
[118] latticeExtra_0.6-30 ggrepel_0.9.3 sass_0.4.7
[121] fastmatch_1.1-4 tools_4.2.0 timechange_0.2.0
[124] parallel_4.2.0 circlize_0.4.15 rstudioapi_0.15.0
[127] foreign_0.8-84 foreach_1.5.2 git2r_0.32.0
[130] gridExtra_2.3 farver_2.1.1 digest_0.6.33
[133] BiocManager_1.30.22 networkD3_0.4 Rcpp_1.0.11
[136] broom_1.0.5 later_1.3.1 httr_1.4.7
[139] ComplexHeatmap_2.12.1 GENIE3_1.18.0 Rdpack_2.5
[142] colorspace_2.1-0 XML_3.99-0.14 fs_1.6.3
[145] splines_4.2.0 statmod_1.5.0 yulab.utils_0.0.8
[148] RBGL_1.72.0 tidytree_0.4.5 graphlayouts_1.0.0
[151] ggplotify_0.1.2 xtable_1.8-4 jsonlite_1.8.7
[154] nloptr_2.0.3 ggtree_3.4.4 tidygraph_1.2.3
[157] ggfun_0.1.2 R6_2.5.1 pillar_1.9.0
[160] htmltools_0.5.6 glue_1.6.2 fastmap_1.1.1
[163] minqa_1.2.5 codetools_0.2-19 fgsea_1.22.0
[166] utf8_1.2.3 sva_3.44.0 lattice_0.21-8
[169] bslib_0.5.1 network_1.18.1 pbkrtest_0.5.2
[172] curl_5.0.2 gtools_3.9.4 interp_1.1-4
[175] survival_3.5-7 statnet.common_4.9.0 rmarkdown_2.24
[178] munsell_0.5.0 GetoptLong_1.0.5 DO.db_2.9
[181] GenomeInfoDbData_1.2.8 iterators_1.0.14 gtable_0.3.4
[184] rbibutils_2.2.15