Johnson_DOX_24_6
Omar Johnson
2024-07-21
Last updated: 2024-07-23
Checks: 7 0
Knit directory: DOX_24_Github/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240723) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 88c6686. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/Johnson_DOX_24_6.html
Ignored: analysis/Johnson_DOX_24_7.html
Ignored: analysis/Johnson_DOX_24_8.html
Ignored: analysis/Johnson_DOX_24_RUV_Limma.html
Untracked files:
Untracked: Fig_2.Rmd
Untracked: Fig_2.html
Untracked: analysis/Johnson_DOX_24_RUV_Limma.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Johnson_DOX_24_6.Rmd) and
HTML (docs/Johnson_DOX_24_6.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 88c6686 | Omar-Johnson | 2024-07-23 | Publish the initial files for myproject |
Load Libraries
Read in Data
Functions
perform_module_comparisons_mutexc_2 <- function(df, module_col, value_col) {
# Ensure the necessary columns exist
if (!(module_col %in% names(df) && value_col %in% names(df))) {
stop("Specified columns do not exist in the dataframe.")
}
# Get a list of all unique modules
modules <- unique(df[[module_col]])
# Initialize an empty list to store combined data frames
combined_df_list <- list()
# Initialize an empty dataframe to store results
results <- data.frame(Module1 = character(),
Module2 = character(),
WilcoxPValue = numeric(),
stringsAsFactors = FALSE)
# Loop through each module
for (module in modules) {
# Data for the current module
current_data <- df %>% filter(!!sym(module_col) == module) %>%
mutate(Group = as.character(module))
# Data for all other modules
other_data <- df %>% filter(!!sym(module_col) != module) %>%
mutate(Group = paste("Not", module, sep=""))
# Combine current module data with other module data
combined_data <- rbind(current_data, other_data)
# Add the combined data to the list
combined_df_list[[module]] <- combined_data
# Perform the Wilcoxon test
test_result <- wilcox.test(current_data[[value_col]], other_data[[value_col]])
# Add the results to the dataframe
results <- rbind(results, data.frame(Module1 = module,
Module2 = "Others",
WilcoxPValue = test_result$p.value))
}
return(list("results" = results, "combined_data" = combined_df_list))
}
# Function assignment
perform_fisher_test_FP <- function(vec1, vec2, vec1_name, vec2_name, plot = FALSE) {
# Create labeled factors for vec1 and vec2
vec1_label <- factor(vec1, labels = c(paste0("Not", vec1_name), paste0("Is", vec1_name)))
vec2_label <- factor(vec2, labels = c(paste0("Not", vec2_name), paste0("Is", vec2_name)))
# Create contingency table with labeled factors
table <- table(vec1_label, vec2_label)
# Perform Fisher's exact test
test_result <- fisher.test(table)
p_value <- test_result$p.value
OR <- test_result$estimate
CI <- test_result$conf.int
# Prepare result
result <- list(
ContingencyTable = table,
PValue = p_value,
Odds_ratio = test_result$estimate,
Confidence_Interval = test_result$conf.int
)
# Generate plot
if (plot) {
# Convert table to data frame for ggplot
table_df <- as.data.frame(as.table(table))
colnames(table_df) <- c("vec1_label", "vec2_label", "Freq")
# Calculate totals for each vec1_label
totals <- aggregate(Freq ~ vec1_label, data = table_df, sum)
# Merge totals with table_df and calculate percentages
table_df <- merge(table_df, totals, by = "vec1_label", all.x = TRUE)
table_df$Percentage <- with(table_df, Freq.x / Freq.y * 100)
table_df$Group <- table_df$vec2_label
# Stacked bar chart
p <- ggplot(table_df, aes(x = vec1_label, y = Percentage, fill = Group)) +
geom_bar(stat = "identity", position = "stack") + # Adjust position to "stack"
facet_wrap(~ vec1_label) +
theme_minimal() +
labs(x = vec1_name, y = "Percentage", fill = vec2_name, title = paste("")) +
theme(axis.text.x = element_text(angle = 45, hjust = 1))
result$Plot <- p
}
return(result)
}
group_by_deciles <- function(x) {
deciles <- cut(x,
breaks = quantile(x, probs = seq(0, 1, by = 0.1), na.rm = TRUE),
include.lowest = TRUE,
labels = paste0("D", 1:10))
return(deciles)
}Fig-6A Triplosensitivity (pTriplo)
pLI_Data <- read.csv(file = GeneDose_File_path, header = TRUE)
pLI_Data %>% dim()[1] 18641 16pLI_Data %>% head() Gene pHaplo pTriplo gnomAD.pLI gnomAD.LOEUF gnomAD.MisOEUF
1 CREBBP 0.999 1.000 1 0.066 0.747
2 MED13L 0.999 1.000 1 0.064 0.741
3 NSD1 0.999 1.000 1 0.095 0.785
4 ARHGEF12 0.999 1.000 1 0.179 0.733
5 CAMTA1 0.999 0.999 1 0.172 0.757
6 CACNA1C 0.999 1.000 1 0.102 0.528
ExAC.DEL.Z.Score ExAC.DUP.Z.Score ExAC.CNV.Z.Score RVIS.Pct. HI.Index sHet
1 1.310 0.848 1.196 0.596 1.000 0.713
2 0.818 -0.319 0.071 1.496 0.722 0.144
3 1.442 0.955 1.314 1.134 0.565 0.237
4 1.640 1.409 1.759 4.633 0.389 0.098
5 0.838 1.395 1.388 7.097 0.196 0.338
6 1.452 -1.872 -1.042 5.767 0.588 0.084
CCDG.DEL.Score CCDG.DUP.Score Episcore EDS
1 0.767 0.934 0.822 0.609
2 0.976 1.103 0.878 0.861
3 0.906 0.856 0.881 0.566
4 0.882 1.174 0.851 0.692
5 1.936 0.807 0.885 0.892
6 1.522 1.518 0.716 0.953pLI_Data_sub <- merge(pLI_Data, New_RNA_PRO_DF_3, by.x = "Gene", by.y = "hgnc_symbol")
pLI_Data_sub_pop <- pLI_Data_sub
# 1. For all genes in genome
ggplot(pLI_Data, aes(x = pTriplo, y=..density..)) +
geom_histogram(position = "dodge", binwidth = 0.01, color = "black") + # Add color for bin borders
theme_minimal() +
labs(title = "pTriplo scores across all genes",
x = "pTriplo score",
y = "Density") Warning: The dot-dot notation (`..density..`) was deprecated in ggplot2 3.4.0.
ℹ Please use `after_stat(density)` instead.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.-1.png)
# 2. For network proteins
ggplot(pLI_Data_sub, aes(x = pTriplo, y=..density..)) +
geom_histogram(position = "dodge", binwidth = 0.01, color = "black") +
theme_minimal() +
labs(title = "pTriplo score for expressed proteins ",
x = "pTriplo score",
y = "Density") -2.png)
# pTriplo Ridge plot
ggplot(pLI_Data_sub, aes(x = pTriplo, y = Modules, fill = Modules)) +
ggridges::geom_density_ridges(alpha = 0.8) +
labs(title = "Dox Associated Module LogFC distribution from DE test",
x = "pTriplo score",
y = "Modules") +
ggridges::theme_ridges() +
scale_fill_identity(guide = "legend")Picking joint bandwidth of 0.0924-3.png)
PlI_mutually_exc <- perform_module_comparisons_mutexc_2(df =pLI_Data_sub , module_col = "Modules", value_col = "pTriplo")
PlI_mutually_exc_combined <- do.call(rbind, PlI_mutually_exc$combined_data)
Group_order <- c("green", "Notgreen","darkgreen", "Notdarkgreen", "midnightblue","Notmidnightblue", "salmon","Notsalmon", "lightyellow", "Notlightyellow", "lightgreen", "Notlightgreen","blue", "Notblue", "magenta" ,"Notmagenta","darkred", "Notdarkred", "brown", "Notbrown", "yellow", "Notyellow","royalblue", "Notroyalblue", "grey", "Notgrey" )
# Factor the Module column in Fulltrait_df
PlI_mutually_exc_combined$Group <- factor(PlI_mutually_exc_combined$Group, levels = Group_order)
PlI_mutually_exc_combined$Group %>% unique() [1] lightyellow Notlightyellow grey Notgrey
[5] royalblue Notroyalblue darkred Notdarkred
[9] darkgreen Notdarkgreen yellow Notyellow
[13] midnightblue Notmidnightblue brown Notbrown
[17] magenta Notmagenta green Notgreen
[21] lightgreen Notlightgreen salmon Notsalmon
[25] blue Notblue
26 Levels: green Notgreen darkgreen Notdarkgreen ... Notgrey# Boxplot
my_colors <- c("green", "green","darkgreen", "darkgreen", "midnightblue","midnightblue", "salmon","salmon", "lightyellow", "lightyellow", "lightgreen", "lightgreen","blue", "blue", "magenta" ,"magenta","darkred", "darkred", "brown", "brown", "yellow", "yellow","royalblue", "royalblue", "grey", "grey")
ggplot(PlI_mutually_exc_combined, aes(x = Group, y = pTriplo, fill = Group)) +
geom_boxplot() +
scale_fill_manual(values = my_colors) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
legend.position = "none") + # Removing the legend
labs(title = "pTriplo",
x = "",
y = "pTriplo score ") +
coord_cartesian(ylim = c(0,1))-4.png)
PlI_mutually_exc$results[PlI_mutually_exc$results$WilcoxPValue < 0.05, ] Module1 Module2 WilcoxPValue
3 royalblue Others 3.453783e-07
10 green Others 2.598900e-17
11 lightgreen Others 4.957535e-02
12 salmon Others 1.702820e-04PlI_mutually_exc_combined_DOX <- PlI_mutually_exc_combined[PlI_mutually_exc_combined$Group %in% c("green", "Notgreen","darkgreen", "Notdarkgreen", "midnightblue","Notmidnightblue", "salmon","Notsalmon", "lightyellow", "Notlightyellow"), ]
ggplot(PlI_mutually_exc_combined_DOX, aes(x = Group, y = pTriplo, fill = Group)) +
geom_boxplot() +
scale_fill_manual(values = my_colors) +
theme_classic() +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
legend.position = "none") + # Removing the legend
labs(title = "pTriplo across DOX-corr. Modules",
x = "",
y = "pTriplo score ") +
coord_cartesian(ylim = c(0,1))-5.png)
Fig-6B Haploinsufficiency (pHaplo)
pLI_Data <- read.csv(file = GeneDose_File_path, header = TRUE)
pLI_Data_sub <- merge(pLI_Data, New_RNA_PRO_DF_3, by.x = "Gene", by.y = "hgnc_symbol")
# 1. For all genes
ggplot(pLI_Data, aes(x = pHaplo, y=..density..)) +
geom_histogram(position = "dodge", binwidth = 0.01, color = "black") + # Add color for bin borders
theme_minimal() +
labs(title = "pHaplo scores across all human genes",
x = "pHaplo score",
y = "Density") -1.png)
# 2. For network proteins
ggplot(pLI_Data_sub, aes(x = pHaplo, y=..density..)) +
geom_histogram(position = "dodge", binwidth = 0.01, color = "black") +
theme_minimal() +
labs(title = "pHaplo score across all genes encoding detected proteins in our data",
x = "pHaplo score",
y = "Density") -2.png)
# pHaplo Ridge plot
ggplot(pLI_Data_sub, aes(x = pHaplo, y = Modules, fill = Modules)) +
ggridges::geom_density_ridges(alpha = 0.8) +
labs(title = "pHaplo scores across modules",
x = "pHaplo score",
y = "") +
ggridges::theme_ridges() +
scale_fill_identity(guide = "legend")Picking joint bandwidth of 0.095-3.png)
PlI_mutually_exc <- perform_module_comparisons_mutexc_2(df =pLI_Data_sub , module_col = "Modules", value_col = "pHaplo")
PlI_mutually_exc_combined <- do.call(rbind, PlI_mutually_exc$combined_data)
Group_order <- c("green", "Notgreen","darkgreen", "Notdarkgreen", "midnightblue","Notmidnightblue", "salmon","Notsalmon", "lightyellow", "Notlightyellow", "lightgreen", "Notlightgreen","blue", "Notblue", "magenta" ,"Notmagenta","darkred", "Notdarkred", "brown", "Notbrown", "yellow", "Notyellow","royalblue", "Notroyalblue", "grey", "Notgrey" )
# Factor the Module column in Fulltrait_df
PlI_mutually_exc_combined$Group <- factor(PlI_mutually_exc_combined$Group, levels = Group_order)
PlI_mutually_exc_combined$Group %>% unique() [1] lightyellow Notlightyellow grey Notgrey
[5] royalblue Notroyalblue darkred Notdarkred
[9] darkgreen Notdarkgreen yellow Notyellow
[13] midnightblue Notmidnightblue brown Notbrown
[17] magenta Notmagenta green Notgreen
[21] lightgreen Notlightgreen salmon Notsalmon
[25] blue Notblue
26 Levels: green Notgreen darkgreen Notdarkgreen ... Notgrey# Boxplot
my_colors <- c("green", "green","darkgreen", "darkgreen", "midnightblue","midnightblue", "salmon","salmon", "lightyellow", "lightyellow", "lightgreen", "lightgreen","blue", "blue", "magenta" ,"magenta","darkred", "darkred", "brown", "brown", "yellow", "yellow","royalblue", "royalblue", "grey", "grey")
ggplot(PlI_mutually_exc_combined, aes(x = Group, y = pHaplo, fill = Group)) +
geom_boxplot() +
scale_fill_manual(values = my_colors) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
legend.position = "none") + # Removing the legend
labs(title = "pHaplo",
x = "",
y = "pHaple score ") +
coord_cartesian(ylim = c(0,1))-4.png)
PlI_mutually_exc$results[PlI_mutually_exc$results$WilcoxPValue < 0.05, ] Module1 Module2 WilcoxPValue
1 lightyellow Others 1.711268e-02
3 royalblue Others 1.037996e-05
10 green Others 9.021108e-21
11 lightgreen Others 4.132253e-02
12 salmon Others 4.035474e-04# Boxplot
my_colors <- c("green", "green","darkgreen", "darkgreen", "midnightblue","midnightblue", "salmon","salmon", "lightyellow", "lightyellow", "lightgreen", "lightgreen","blue", "blue", "magenta" ,"magenta","darkred", "darkred", "brown", "brown", "yellow", "yellow","royalblue", "royalblue", "grey", "grey")
PlI_mutually_exc_combined_DOX <- PlI_mutually_exc_combined[PlI_mutually_exc_combined$Group %in% c("green", "Notgreen","darkgreen", "Notdarkgreen", "midnightblue","Notmidnightblue", "salmon","Notsalmon", "lightyellow", "Notlightyellow"), ]
ggplot(PlI_mutually_exc_combined_DOX, aes(x = Group, y = pHaplo, fill = Group)) +
geom_boxplot() +
scale_fill_manual(values = my_colors) +
theme_classic() +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
legend.position = "none") + # Removing the legend
labs(title = "pHaplo accross DOX-corr. Modules",
x = "",
y = "pHaple score ") +
coord_cartesian(ylim = c(0,1))-5.png)
PlI_mutually_exc$results[PlI_mutually_exc$results$WilcoxPValue < 0.05, ] Module1 Module2 WilcoxPValue
1 lightyellow Others 1.711268e-02
3 royalblue Others 1.037996e-05
10 green Others 9.021108e-21
11 lightgreen Others 4.132253e-02
12 salmon Others 4.035474e-04Fig-6C Probability loss of function intolerant (pLI)
pLI_Data <- read.csv(file = pLI_DF_File_path, header = TRUE)
pLI_Data %>% dim()[1] 19704 2pLI_Data_sub <- merge(pLI_Data, New_RNA_PRO_DF_3, by.x = "gene", by.y = "hgnc_symbol")
pLI_Data_sub_pop <- pLI_Data_sub
pLI_Data_sub_pop_hub <- pLI_Data_sub_pop[pLI_Data_sub_pop$Protein %in% hubs$Gene, ]
# 1. For all genes
ggplot(pLI_Data, aes(x = pLI, y=..density..)) +
geom_histogram(position = "dodge", binwidth = 0.1) +
theme_minimal() +
labs(title = "pLI scores aacross all human genes",
x = "pLI Score",
y = "density") Warning: Removed 507 rows containing non-finite values (`stat_bin()`).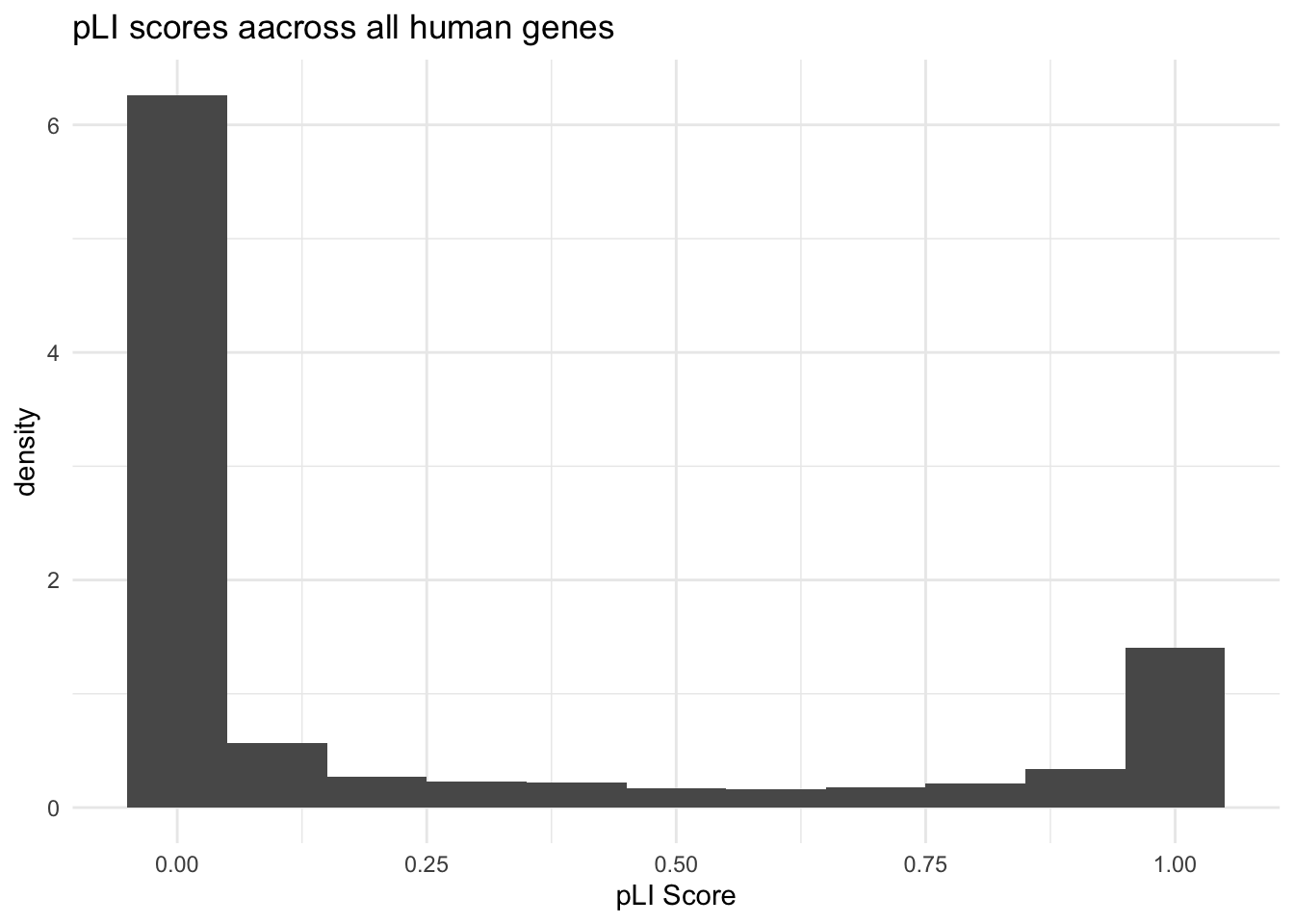
# 2. For network proteins
ggplot(pLI_Data_sub, aes(x = pLI, y=..density..)) +
geom_histogram(position = "dodge", binwidth = 0.1) +
theme_minimal() +
labs(title = "pLI scores across all detected iPSC-CM proteins ",
x = "pLI Score",
y = "density") Warning: Removed 1 rows containing non-finite values (`stat_bin()`).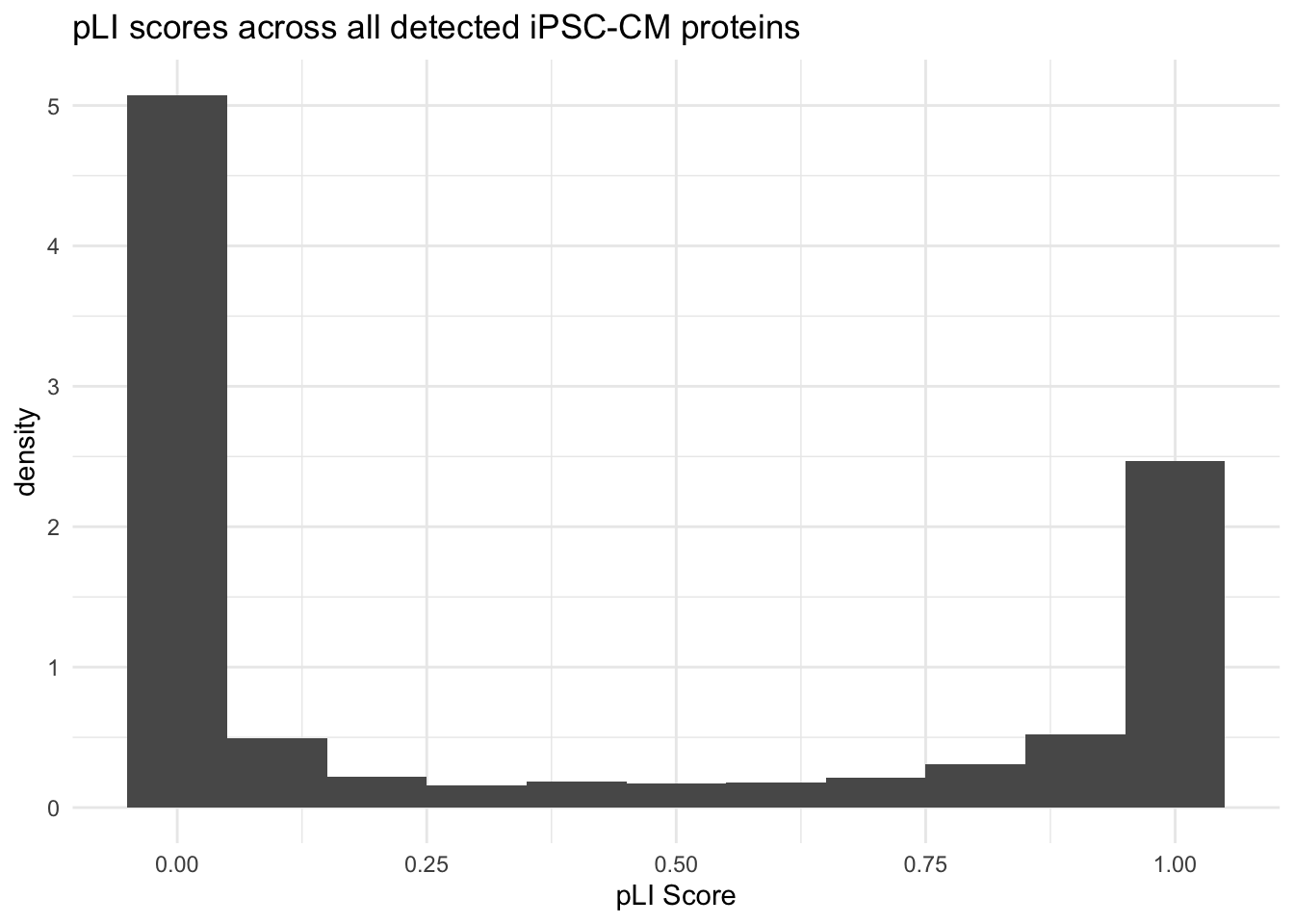
# 1. For all detected proteins across modules
ggplot(pLI_Data_sub, aes(x = pLI, fill = Modules, y=..density..)) +
geom_histogram(position = "dodge", binwidth = 0.2) +
theme_minimal() +
labs(title = "pLI scores for all detected proteins across modules",
x = "pLI Score",
y = "density") +
scale_fill_identity(guide = "legend")Warning: Removed 1 rows containing non-finite values (`stat_bin()`).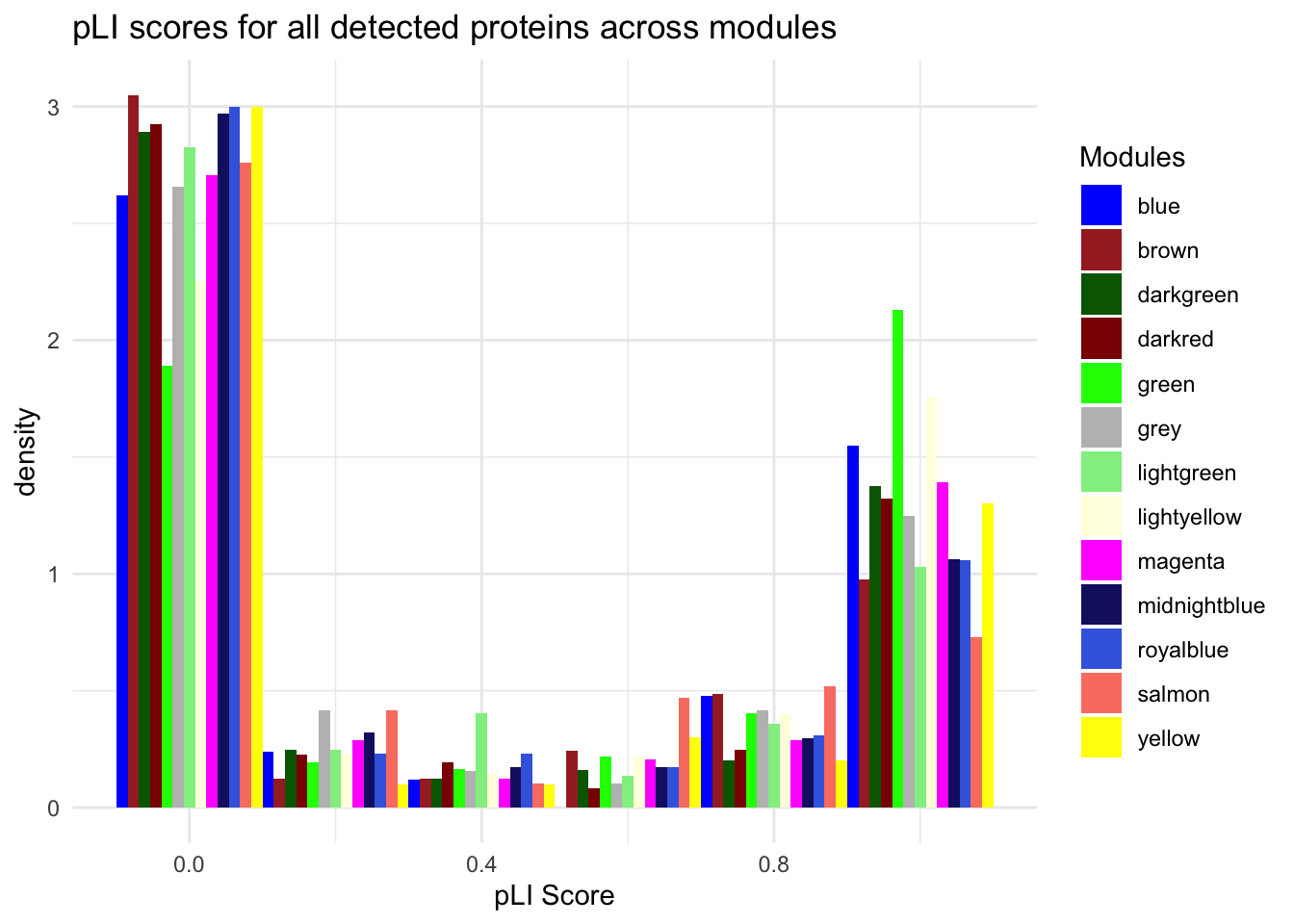
ggplot(pLI_Data_sub, aes(x = Modules, y = pLI, fill = Modules)) +
geom_boxplot() +
geom_jitter(width = 0.1, size = 1, alpha = 0.2, col = "black") + # Add this line
scale_fill_identity() +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
labs(title = "pLI scores across modules",
x = "Module",
y = "pLI Score") +
scale_fill_identity(guide = "legend")Scale for fill is already present.
Adding another scale for fill, which will replace the existing scale.Warning: Removed 1 rows containing non-finite values (`stat_boxplot()`).Warning: Removed 1 rows containing missing values (`geom_point()`).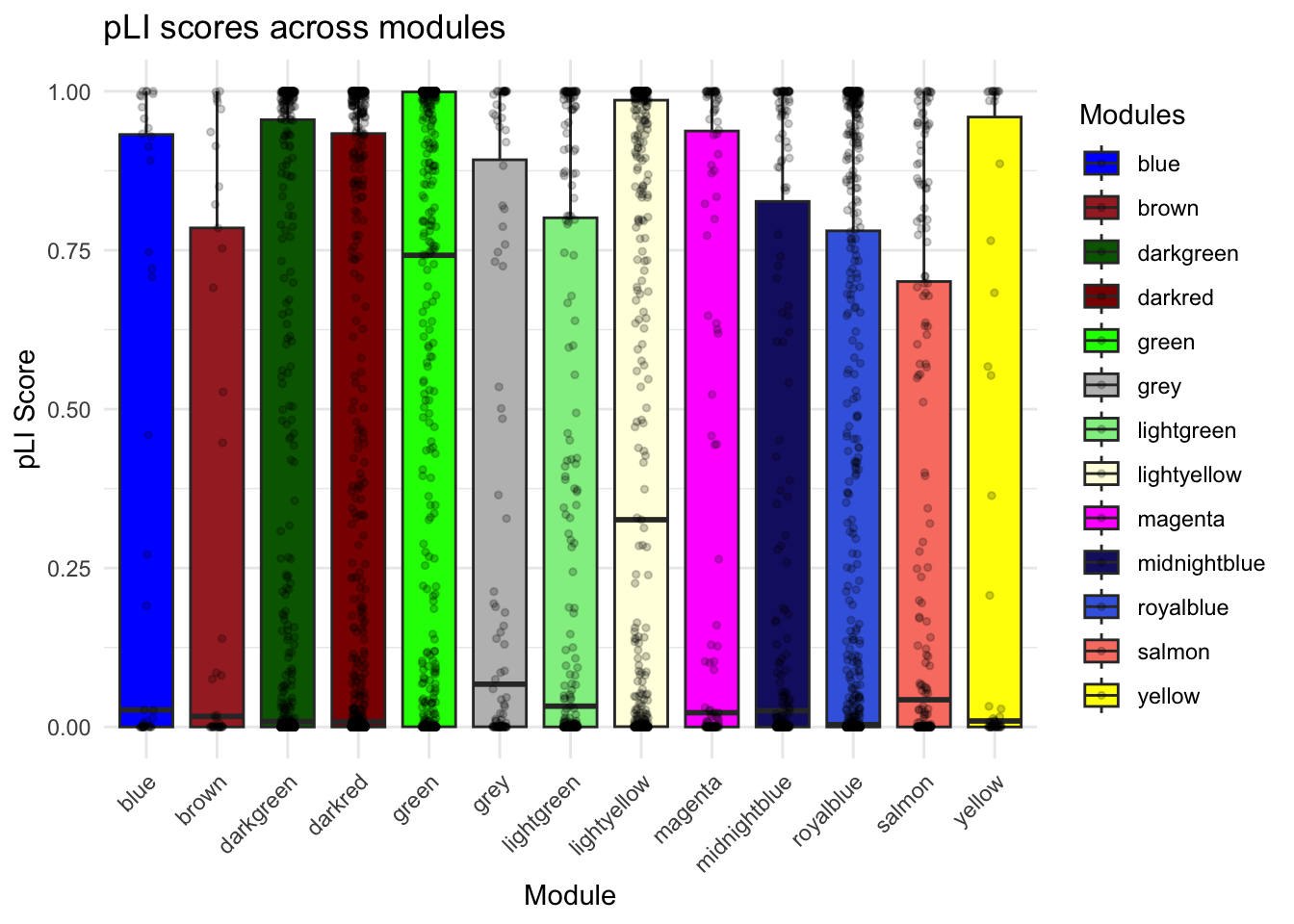
PlI_mutually_exc <- perform_module_comparisons_mutexc_2(df = pLI_Data_sub , module_col = "Modules", value_col = "pLI")
PlI_mutually_exc_combined <- do.call(rbind, PlI_mutually_exc$combined_data)
Group_order <- c("green", "Notgreen","darkgreen", "Notdarkgreen", "midnightblue","Notmidnightblue", "salmon","Notsalmon", "lightyellow", "Notlightyellow", "lightgreen", "Notlightgreen","blue", "Notblue", "magenta" ,"Notmagenta","darkred", "Notdarkred", "brown", "Notbrown", "yellow", "Notyellow","royalblue", "Notroyalblue", "grey", "Notgrey" )
# Factor the Module column in Fulltrait_df
PlI_mutually_exc_combined$Group <- factor(PlI_mutually_exc_combined$Group, levels = Group_order)
PlI_mutually_exc_combined$Group %>% unique() [1] lightyellow Notlightyellow grey Notgrey
[5] royalblue Notroyalblue darkred Notdarkred
[9] darkgreen Notdarkgreen yellow Notyellow
[13] midnightblue Notmidnightblue brown Notbrown
[17] magenta Notmagenta green Notgreen
[21] lightgreen Notlightgreen salmon Notsalmon
[25] blue Notblue
26 Levels: green Notgreen darkgreen Notdarkgreen ... Notgrey# Boxplot
my_colors <- c("green", "green","darkgreen", "darkgreen", "midnightblue","midnightblue", "salmon","salmon", "lightyellow", "lightyellow", "lightgreen", "lightgreen","blue", "blue", "magenta" ,"magenta","darkred", "darkred", "brown", "brown", "yellow", "yellow","royalblue", "royalblue", "grey", "grey")
ggplot(PlI_mutually_exc_combined, aes(x = Group, y = pLI, fill = Group)) +
geom_boxplot() +
scale_fill_manual(values = my_colors) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
legend.position = "none") + # Removing the legend
labs(title = "probability of being loss-of-function intolerant",
x = "",
y = "PLI score ") +
ylim(0, 1)Warning: Removed 13 rows containing non-finite values (`stat_boxplot()`).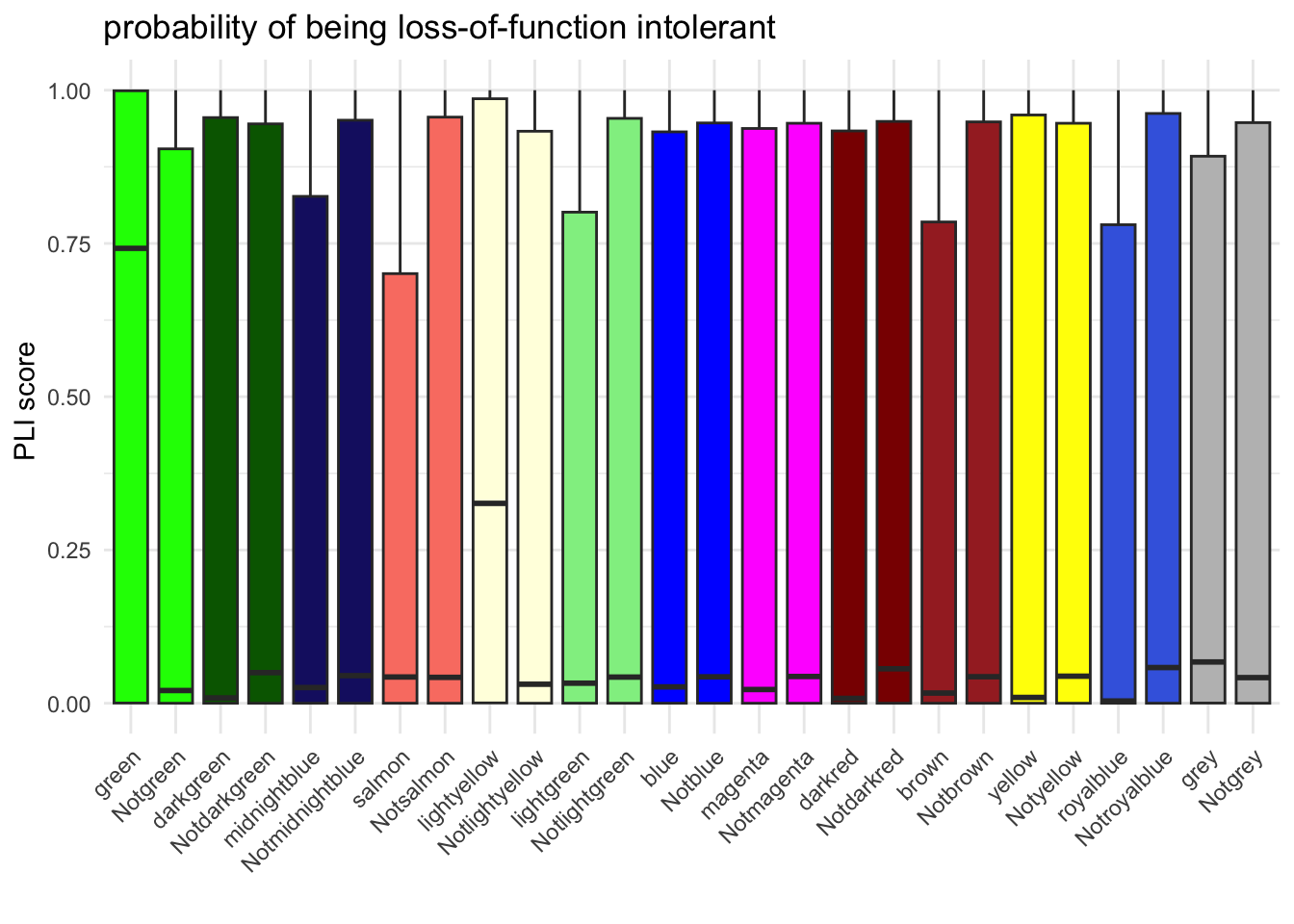
PlI_mutually_exc$results[PlI_mutually_exc$results$WilcoxPValue < 0.05, ] Module1 Module2 WilcoxPValue
1 lightyellow Others 4.798651e-07
3 royalblue Others 1.222661e-08
4 darkred Others 1.483659e-03
5 darkgreen Others 3.993146e-02
10 green Others 2.618431e-19PlI_mutually_exc_combined_DOX <- PlI_mutually_exc_combined[PlI_mutually_exc_combined$Group %in% c("green", "Notgreen","darkgreen", "Notdarkgreen", "midnightblue","Notmidnightblue", "salmon","Notsalmon", "lightyellow", "Notlightyellow"), ]
ggplot(PlI_mutually_exc_combined_DOX, aes(x = Group, y = pLI, fill = Group)) +
geom_boxplot() +
scale_fill_manual(values = my_colors) +
theme_classic() +
theme(axis.text.x = element_text(angle = 45, hjust = 1),
legend.position = "none") + # Removing the legend
labs(title = "pLI across DOX-corr. modules",
x = "",
y = "PLI score ") +
ylim(0, 1)Warning: Removed 5 rows containing non-finite values (`stat_boxplot()`).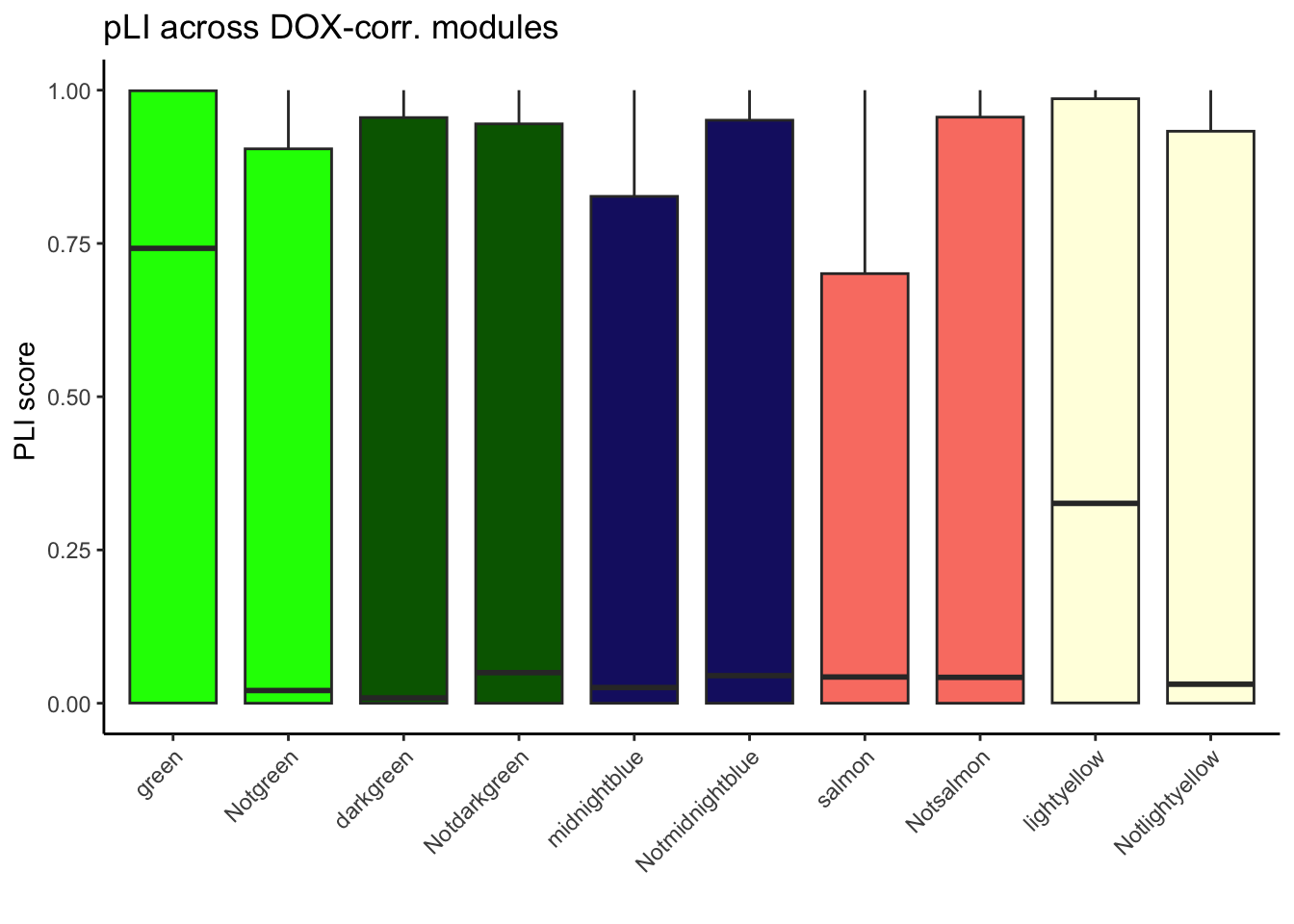
PlI_mutually_exc$results[PlI_mutually_exc$results$WilcoxPValue < 0.05, ] Module1 Module2 WilcoxPValue
1 lightyellow Others 4.798651e-07
3 royalblue Others 1.222661e-08
4 darkred Others 1.483659e-03
5 darkgreen Others 3.993146e-02
10 green Others 2.618431e-19Fig-6D pLI & Norm_K
pLI_kIN_DF_TT <- merge(Toptable_Modules, pLI_Data_sub, by.x = "Protein" , by.y = "Protein")
pLI_kIN_DF_TT %>% colnames() [1] "Protein" "X.x" "kTotal.x"
[4] "kWithin.x" "kOut.x" "kDiff.x"
[7] "logFC.x" "AveExpr.x" "t.x"
[10] "P.Value.x" "adj.P.Val.x" "B.x"
[13] "threshold_P.x" "Modules.x" "DE_or_Not.x"
[16] "Norm_kIN.x" "Norm_kOut.x" "logFC.y.x"
[19] "AveExpr.y.x" "t.y.x" "P.Value.y.x"
[22] "adj.P.Val.y.x" "B.y.x" "threshold_P.y.x"
[25] "Modules.y.x" "DE_or_Not.y.x" "Is_DA.x"
[28] "Is_DOXcorrelated.x" "Is_Hub.x" "Is_Cis_pQTL.x"
[31] "Is_Trans_pQTL.x" "Is_pQTL.x" "pLI_assigned.x"
[34] "pLI_Mut.Intolerant.x" "pLI_Mut.Tolerant.x" "Is_Druggable.x"
[37] "Is_CVD_protein.x" "Is_CVD_PPI_protein.x" "gene"
[40] "pLI" "X.y" "kTotal.y"
[43] "kWithin.y" "kOut.y" "kDiff.y"
[46] "logFC.y" "AveExpr.y" "t.y"
[49] "P.Value.y" "adj.P.Val.y" "B.y"
[52] "threshold_P.y" "Modules.y" "DE_or_Not.y"
[55] "Norm_kIN.y" "Norm_kOut.y" "logFC.y.y"
[58] "AveExpr.y.y" "t.y.y" "P.Value.y.y"
[61] "adj.P.Val.y.y" "B.y.y" "threshold_P.y.y"
[64] "Modules.y.y" "DE_or_Not.y.y" "Is_DA.y"
[67] "Is_DOXcorrelated.y" "Is_Hub.y" "Is_Cis_pQTL.y"
[70] "Is_Trans_pQTL.y" "Is_pQTL.y" "pLI_assigned.y"
[73] "pLI_Mut.Intolerant.y" "pLI_Mut.Tolerant.y" "Is_Druggable.y"
[76] "Is_CVD_protein.y" "Is_CVD_PPI_protein.y" "RNA_LogFC"
[79] "RNA_Pval" "Pro_LogFC" "Pro_Pval"
[82] "sign_match" pLI_kIN_DF_TT_2 <- pLI_kIN_DF_TT[,c(1,2,3,4,5,6,7,10,14,16,17,28,33,34,35,38,40)]
pLI_kIN_DF_TT_2 %>% colnames() [1] "Protein" "X.x" "kTotal.x"
[4] "kWithin.x" "kOut.x" "kDiff.x"
[7] "logFC.x" "P.Value.x" "Modules.x"
[10] "Norm_kIN.x" "Norm_kOut.x" "Is_DOXcorrelated.x"
[13] "pLI_assigned.x" "pLI_Mut.Intolerant.x" "pLI_Mut.Tolerant.x"
[16] "Is_CVD_PPI_protein.x" "pLI" colnames(pLI_kIN_DF_TT_2) <- c("Protein","X","kTotal","kWithin","kOut", "kDiff", "logFC", "P.Value", "Modules", "Norm_kIN", "Norm_kOut","Is_DOXcorrelated", "pLI_assigned","pLI_Mut.Intolerant","pLI_Mut.Tolerant", "Is_CVD_PPI_protein", "pLI")
pLI_kIN_DF_TT_unique <- pLI_kIN_DF_TT_2 %>%
distinct(Protein, .keep_all = TRUE)
# Calculate the total number of members
total_net_members <- nrow(pLI_kIN_DF_TT_unique)
total_net_members[1] 3846# Normalize kOut by the number of members not in the module
pLI_kIN_DF_TT_unique_2 <- pLI_kIN_DF_TT_unique %>%
group_by(Modules) %>%
mutate(Norm_kIN = kWithin / n(),
Norm_kOut = kOut / (total_net_members - n())) %>%
ungroup()
pLI_kIN_DF_TT_unique_2$Norm_kOut %>% hist(breaks = 20)
pLI_kIN_DF_TT_unique_2_high <- pLI_kIN_DF_TT_unique_2[pLI_kIN_DF_TT_unique_2$pLI >= 0.9, ]
pLI_kIN_DF_TT_unique_2_high %>% dim()[1] 1067 17pLI_kIN_DF_TT_unique_2_low <- pLI_kIN_DF_TT_unique_2[pLI_kIN_DF_TT_unique_2$pLI <= 0.1, ]
pLI_kIN_DF_TT_unique_2_low %>% dim()[1] 2066 17wilcox.test(pLI_kIN_DF_TT_unique_2_high$Norm_kIN, pLI_kIN_DF_TT_unique_2_low$Norm_kIN)
Wilcoxon rank sum test with continuity correction
data: pLI_kIN_DF_TT_unique_2_high$Norm_kIN and pLI_kIN_DF_TT_unique_2_low$Norm_kIN
W = 1235856, p-value = 1.691e-08
alternative hypothesis: true location shift is not equal to 0wilcox.test(pLI_kIN_DF_TT_unique_2_high$Norm_kOut, pLI_kIN_DF_TT_unique_2_low$Norm_kOut)
Wilcoxon rank sum test with continuity correction
data: pLI_kIN_DF_TT_unique_2_high$Norm_kOut and pLI_kIN_DF_TT_unique_2_low$Norm_kOut
W = 1203212, p-value = 1.877e-05
alternative hypothesis: true location shift is not equal to 0# Create a data frame
pLI_Hubs_DF_Boxplot <- data.frame(
# 1. Generate values to compare
values = c(pLI_kIN_DF_TT_unique_2_high$Norm_kIN,
pLI_kIN_DF_TT_unique_2_low$Norm_kIN),
# 2. Factor values to compare
group = factor(c(rep("High_pLI_kIN", length(pLI_kIN_DF_TT_unique_2_high$Norm_kIN)),
c(rep("Low_pLI_kIN", length(pLI_kIN_DF_TT_unique_2_low$Norm_kIN)))
))
)
# Create boxplot
ggplot(pLI_Hubs_DF_Boxplot, aes(x = group, y = values)) +
geom_boxplot() +
labs(x = "Group", y = "Normalized intramodular connectivity (kIN)") +
ggtitle("Connectivity differences between mutatiion tolerant/intolerant proteins")+
theme_classic()+
coord_cartesian(ylim = c(0, 0.25))Warning: Removed 2 rows containing non-finite values (`stat_boxplot()`).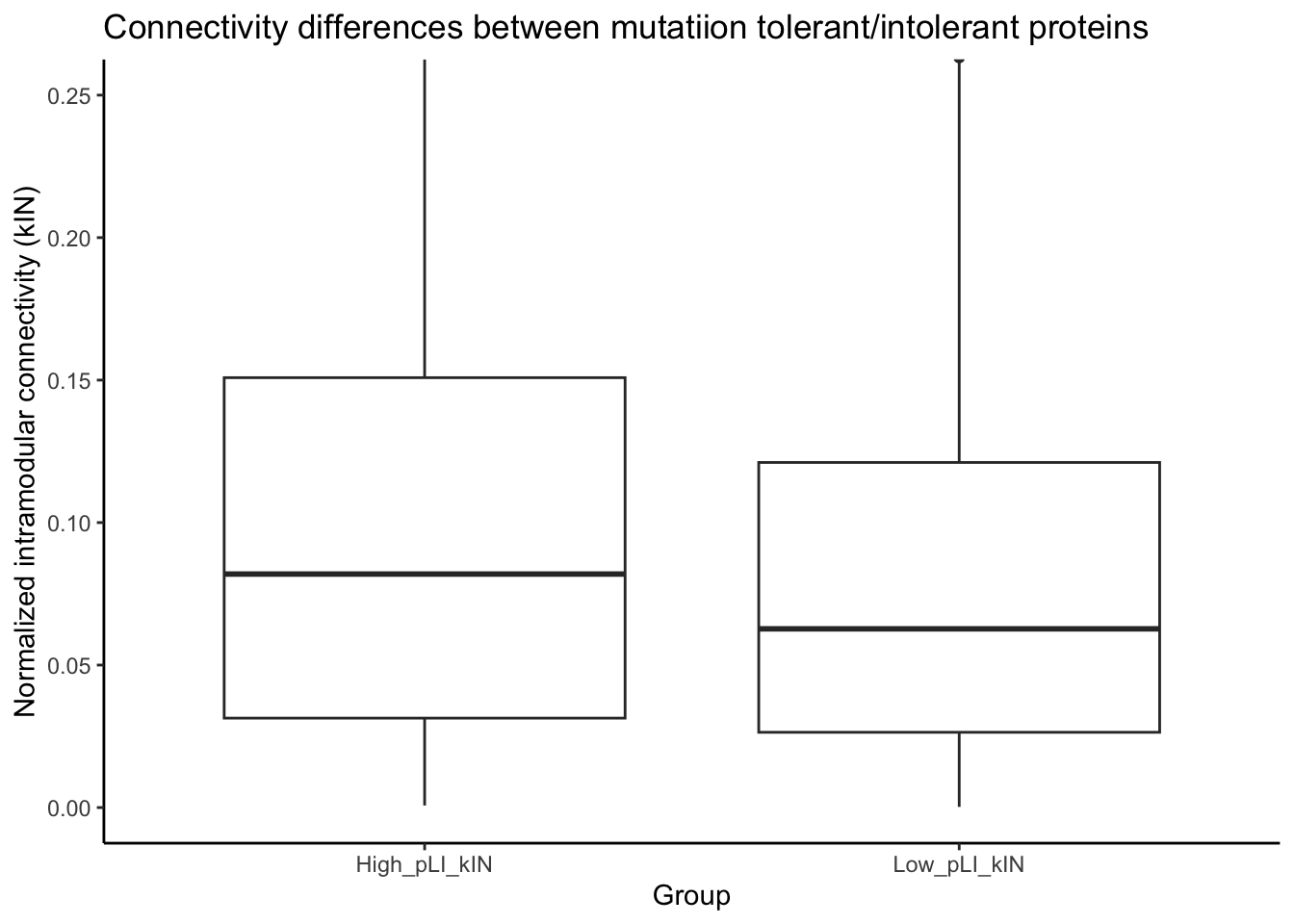
Fig-6E pLI & all connectivity
pLI_kIN_DF_TT_unique$DOXcorchar <- as.character(pLI_kIN_DF_TT_unique$Is_DOXcorrelated)
pLI_data_summary_KIN_Tot <- pLI_kIN_DF_TT_unique
pLI_data_summary_KIN_NormKIN <- pLI_kIN_DF_TT_unique
pLI_data_summary_KIN_NormkOUT <- pLI_kIN_DF_TT_unique
pLI_data_summary_KIN_Tot$Decile <-
group_by_deciles(pLI_data_summary_KIN_Tot$kTotal)
pLI_data_summary_KIN_Tot$Type <- c("kTotal")
pLI_data_summary_KIN_NormKIN$Decile <-
group_by_deciles(pLI_data_summary_KIN_NormKIN$Norm_kIN)
pLI_data_summary_KIN_NormKIN$Type <- c("kIN")
pLI_data_summary_KIN_NormkOUT$Decile <-
group_by_deciles(pLI_data_summary_KIN_NormkOUT$Norm_kOut)
pLI_data_summary_KIN_NormkOUT$Type <- c("kOut")
pLI_data_summary_KIN_ALL <- rbind(pLI_data_summary_KIN_Tot,pLI_data_summary_KIN_NormKIN,pLI_data_summary_KIN_NormkOUT)
pLI_data_summary_KIN_ALL %>% colnames() [1] "Protein" "X" "kTotal"
[4] "kWithin" "kOut" "kDiff"
[7] "logFC" "P.Value" "Modules"
[10] "Norm_kIN" "Norm_kOut" "Is_DOXcorrelated"
[13] "pLI_assigned" "pLI_Mut.Intolerant" "pLI_Mut.Tolerant"
[16] "Is_CVD_PPI_protein" "pLI" "DOXcorchar"
[19] "Decile" "Type" # Summarize the data
pLI_connectivity_summarized <- pLI_data_summary_KIN_ALL %>%
group_by(Type, DOXcorchar, Decile) %>%
summarize(median_pLI = median(pLI, na.rm = TRUE)) %>%
ungroup()`summarise()` has grouped output by 'Type', 'DOXcorchar'. You can override
using the `.groups` argument.pLI_connectivity_summarized$Type <- factor(x = pLI_connectivity_summarized$Type, levels = c("kTotal","kIN","kOut"))
ggplot(pLI_connectivity_summarized, aes(x = Decile, y = median_pLI, color = DOXcorchar, linetype = Type, group = interaction(Type, DOXcorchar))) +
geom_line(size = 1) +
scale_y_continuous(breaks = seq(0, 1, by = 0.1), limits = c(0, 1)) + # Add y-axis ticks
labs(title = "Mutation intolerance determined by connectivity",
x = "Connetivity Deciles",
y = "Probabilty of mutation intolerance (pLI)",
color = "DOXcorchar",
linetype = "Type") +
theme_classic()Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.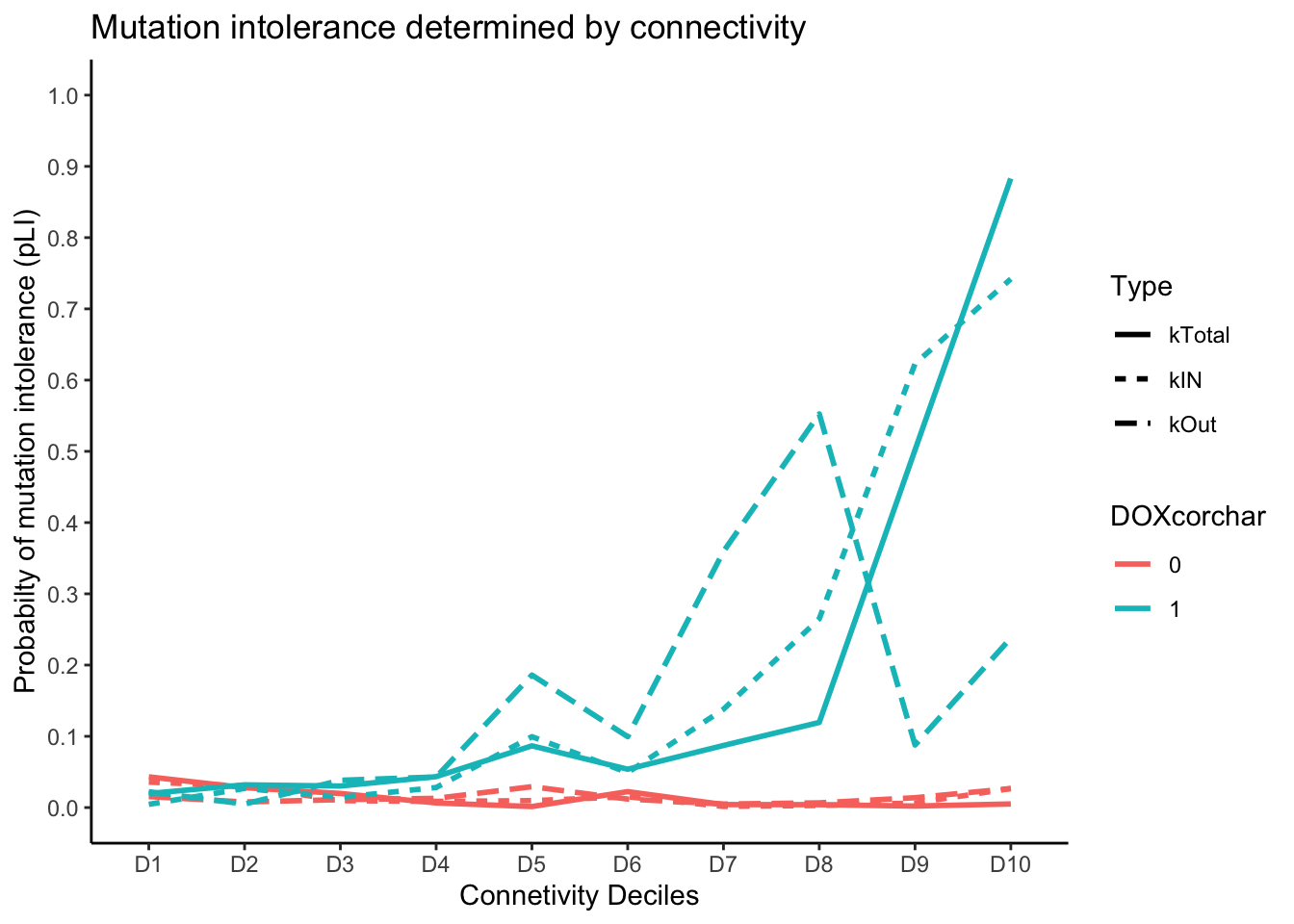
fig-6F Mutation tolerant/intolerant enrichment
pLI_Data <- read.csv(file = pLI_DF_File_path, header = TRUE)
pLI_Data_sub <- merge(pLI_Data, New_RNA_PRO_DF_3, by.x = "gene", by.y = "hgnc_symbol")
pLI_Data_sub2 <- pLI_Data_sub
pLI_Data_sub2 <- pLI_Data_sub2 %>%
mutate(Hub = if_else(pLI_Data_sub2$Protein %in% hubs$Gene, 1, 0))
pLI_Data_sub2 <- pLI_Data_sub2 %>%
mutate(DOXcorr = if_else(pLI_Data_sub2$Modules %in% c("green","darkgreen", "midnightblue", "salmon","lightyellow"), 1, 0))
pLI_Data_sub2 <- pLI_Data_sub2 %>%
mutate(Mut_Intolerant = if_else(pLI_Data_sub2$pLI >= 0.9, 1, 0))
pLI_Data_sub2 <- pLI_Data_sub2 %>%
mutate(Mut_Resistant = if_else(pLI_Data_sub2$pLI <= 0.1 , 1, 0))
pLI_Data_sub2_hub <- pLI_Data_sub2[pLI_Data_sub2$Hub == 1, ]
# Tests
FP_DOXcorr_High <- perform_fisher_test_FP(vec1 = pLI_Data_sub2$DOXcorr, vec2 = pLI_Data_sub2$Mut_Intolerant, vec1_name = "", vec2_name = "", plot = FALSE)
FP_DOXcorr_Low <- perform_fisher_test_FP(vec1 = pLI_Data_sub2$DOXcorr, vec2 = pLI_Data_sub2$Mut_Resistant, vec1_name = "", vec2_name = "", plot = FALSE)
FP_Hub_High <- perform_fisher_test_FP(vec1 = pLI_Data_sub2$Hub, vec2 = pLI_Data_sub2$Mut_Intolerant, vec1_name = "", vec2_name = "", plot = FALSE)
FP_Hub_Low <- perform_fisher_test_FP(vec1 = pLI_Data_sub2$Hub, vec2 = pLI_Data_sub2$Mut_Resistant, vec1_name = "", vec2_name = "", plot = FALSE)
FP_DOXcorr_Hub_High <- perform_fisher_test_FP(vec1 = pLI_Data_sub2_hub$DOXcorr, vec2 = pLI_Data_sub2_hub$Mut_Intolerant, vec1_name = "", vec2_name = "", plot = FALSE)
FP_DOXcorr_Hub_Low <- perform_fisher_test_FP(vec1 = pLI_Data_sub2_hub$DOXcorr, vec2 = pLI_Data_sub2_hub$Mut_Resistant, vec1_name = "", vec2_name = "", plot = FALSE)
FP_List <- list(FP_DOXcorr_High, FP_DOXcorr_Low, FP_Hub_High, FP_Hub_Low, FP_DOXcorr_Hub_High, FP_DOXcorr_Hub_Low)
FP_DF <- data.frame(
Odds_ratio = numeric(length(FP_List)),
Lower_CI = numeric(length(FP_List)),
Upper_CI = numeric(length(FP_List)),
Pval = numeric(length(FP_List))
)
for (i in 1:length(FP_List)) {
FP_DF$Odds_ratio[i] <- FP_List[[i]]$Odds_ratio
FP_DF$Lower_CI[i] <- FP_List[[i]]$Confidence_Interval[1]
FP_DF$Upper_CI[i] <- FP_List[[i]]$Confidence_Interval[2]
FP_DF$Pval[i] <- FP_List[[i]]$PValue
}
# Add row names for the labels in the forest plot
FP_DF$Label <- c("FP_DOXcorr_High", "FP_DOXcorr_Low", "FP_Hub_High", "FP_Hub_Low", "FP_DOXcorr_Hub_High", "FP_DOXcorr_Hub_Low")
FP_DF$Label <- factor(FP_DF$Label, levels = rev(c("FP_Hub_High", "FP_Hub_Low", "FP_DOXcorr_High", "FP_DOXcorr_Low", "FP_DOXcorr_Hub_High", "FP_DOXcorr_Hub_Low")))
# Add a new column to indicate "cis" or "trans"
FP_DF$Type <- ifelse(grepl("High", FP_DF$Label), "Mutation Intolerant", "Mutation Tolerant")
# Plot with color based on "Type"
ggplot(FP_DF, aes(x = Label, y = Odds_ratio, ymin = Lower_CI, ymax = Upper_CI, color = Type)) +
geom_pointrange() +
geom_hline(yintercept = 1, linetype = "dashed", color = "red") +
coord_flip() +
labs(
title = "Odds of mutation tolerance",
x = "",
y = "Odds Ratio (95% CI)"
) +
theme_classic() +
scale_color_manual(values = c("Mutation Tolerant" = "dodgerblue4", "Mutation Intolerant" = "darkred"))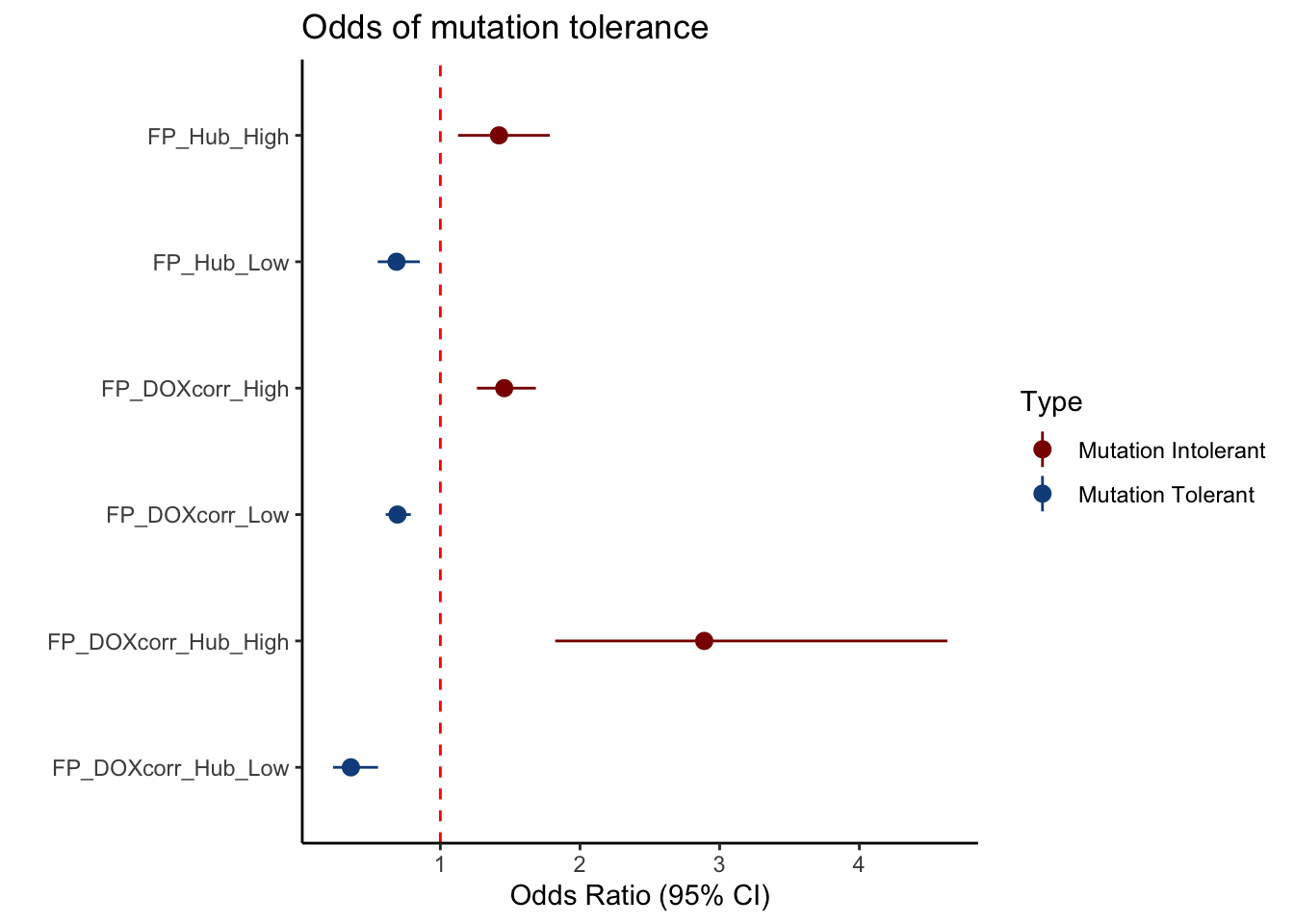
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur/Monterey 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] scales_1.2.1
[2] ReactomePA_1.40.0
[3] impute_1.70.0
[4] WGCNA_1.72-1
[5] fastcluster_1.2.3
[6] dynamicTreeCut_1.63-1
[7] BioNERO_1.4.2
[8] reshape2_1.4.4
[9] ggridges_0.5.4
[10] biomaRt_2.52.0
[11] ggvenn_0.1.10
[12] UpSetR_1.4.0
[13] DOSE_3.22.1
[14] variancePartition_1.26.0
[15] clusterProfiler_4.4.4
[16] pheatmap_1.0.12
[17] qvalue_2.28.0
[18] Homo.sapiens_1.3.1
[19] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[20] org.Hs.eg.db_3.15.0
[21] GO.db_3.15.0
[22] OrganismDbi_1.38.1
[23] GenomicFeatures_1.48.4
[24] AnnotationDbi_1.58.0
[25] cluster_2.1.4
[26] ggfortify_0.4.16
[27] lubridate_1.9.2
[28] forcats_1.0.0
[29] stringr_1.5.0
[30] dplyr_1.1.2
[31] purrr_1.0.2
[32] readr_2.1.4
[33] tidyr_1.3.0
[34] tibble_3.2.1
[35] ggplot2_3.4.3
[36] tidyverse_2.0.0
[37] RColorBrewer_1.1-3
[38] RUVSeq_1.30.0
[39] edgeR_3.38.4
[40] limma_3.52.4
[41] EDASeq_2.30.0
[42] ShortRead_1.54.0
[43] GenomicAlignments_1.32.1
[44] SummarizedExperiment_1.26.1
[45] MatrixGenerics_1.8.1
[46] matrixStats_1.0.0
[47] Rsamtools_2.12.0
[48] GenomicRanges_1.48.0
[49] Biostrings_2.64.1
[50] GenomeInfoDb_1.32.4
[51] XVector_0.36.0
[52] IRanges_2.30.1
[53] S4Vectors_0.34.0
[54] BiocParallel_1.30.4
[55] Biobase_2.56.0
[56] BiocGenerics_0.42.0
[57] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] rappdirs_0.3.3 rtracklayer_1.56.1 minet_3.54.0
[4] R.methodsS3_1.8.2 coda_0.19-4 bit64_4.0.5
[7] knitr_1.43 aroma.light_3.26.0 DelayedArray_0.22.0
[10] R.utils_2.12.2 rpart_4.1.19 data.table_1.14.8
[13] hwriter_1.3.2.1 KEGGREST_1.36.3 RCurl_1.98-1.12
[16] doParallel_1.0.17 generics_0.1.3 preprocessCore_1.58.0
[19] callr_3.7.3 RhpcBLASctl_0.23-42 RSQLite_2.3.1
[22] shadowtext_0.1.2 bit_4.0.5 tzdb_0.4.0
[25] enrichplot_1.16.2 xml2_1.3.5 httpuv_1.6.11
[28] viridis_0.6.4 xfun_0.40 hms_1.1.3
[31] jquerylib_0.1.4 evaluate_0.21 promises_1.2.1
[34] fansi_1.0.4 restfulr_0.0.15 progress_1.2.2
[37] caTools_1.18.2 dbplyr_2.3.3 htmlwidgets_1.6.2
[40] igraph_1.5.1 DBI_1.1.3 ggnewscale_0.4.9
[43] backports_1.4.1 annotate_1.74.0 aod_1.3.2
[46] deldir_1.0-9 vctrs_0.6.3 abind_1.4-5
[49] cachem_1.0.8 withr_2.5.0 ggforce_0.4.1
[52] checkmate_2.2.0 treeio_1.20.2 prettyunits_1.1.1
[55] ape_5.7-1 lazyeval_0.2.2 crayon_1.5.2
[58] genefilter_1.78.0 labeling_0.4.2 pkgconfig_2.0.3
[61] tweenr_2.0.2 nlme_3.1-163 nnet_7.3-19
[64] rlang_1.1.1 lifecycle_1.0.3 downloader_0.4
[67] filelock_1.0.2 BiocFileCache_2.4.0 rprojroot_2.0.3
[70] polyclip_1.10-4 graph_1.74.0 Matrix_1.5-4.1
[73] aplot_0.2.0 NetRep_1.2.7 boot_1.3-28.1
[76] base64enc_0.1-3 GlobalOptions_0.1.2 whisker_0.4.1
[79] processx_3.8.2 png_0.1-8 viridisLite_0.4.2
[82] rjson_0.2.21 bitops_1.0-7 getPass_0.2-2
[85] R.oo_1.25.0 ggnetwork_0.5.12 KernSmooth_2.23-22
[88] blob_1.2.4 shape_1.4.6 jpeg_0.1-10
[91] gridGraphics_0.5-1 reactome.db_1.81.0 graphite_1.42.0
[94] memoise_2.0.1 magrittr_2.0.3 plyr_1.8.8
[97] gplots_3.1.3 zlibbioc_1.42.0 compiler_4.2.0
[100] scatterpie_0.2.1 BiocIO_1.6.0 clue_0.3-64
[103] intergraph_2.0-3 lme4_1.1-34 cli_3.6.1
[106] patchwork_1.1.3 ps_1.7.5 htmlTable_2.4.1
[109] Formula_1.2-5 mgcv_1.9-0 MASS_7.3-60
[112] tidyselect_1.2.0 stringi_1.7.12 highr_0.10
[115] yaml_2.3.7 GOSemSim_2.22.0 locfit_1.5-9.8
[118] latticeExtra_0.6-30 ggrepel_0.9.3 sass_0.4.7
[121] fastmatch_1.1-4 tools_4.2.0 timechange_0.2.0
[124] parallel_4.2.0 circlize_0.4.15 rstudioapi_0.15.0
[127] foreign_0.8-84 foreach_1.5.2 git2r_0.32.0
[130] gridExtra_2.3 farver_2.1.1 ggraph_2.1.0
[133] digest_0.6.33 BiocManager_1.30.22 networkD3_0.4
[136] Rcpp_1.0.11 broom_1.0.5 later_1.3.1
[139] httr_1.4.7 ComplexHeatmap_2.12.1 GENIE3_1.18.0
[142] Rdpack_2.5 colorspace_2.1-0 XML_3.99-0.14
[145] fs_1.6.3 splines_4.2.0 statmod_1.5.0
[148] yulab.utils_0.0.8 RBGL_1.72.0 tidytree_0.4.5
[151] graphlayouts_1.0.0 ggplotify_0.1.2 xtable_1.8-4
[154] jsonlite_1.8.7 nloptr_2.0.3 ggtree_3.4.4
[157] tidygraph_1.2.3 ggfun_0.1.2 R6_2.5.1
[160] Hmisc_5.1-0 pillar_1.9.0 htmltools_0.5.6
[163] glue_1.6.2 fastmap_1.1.1 minqa_1.2.5
[166] codetools_0.2-19 fgsea_1.22.0 utf8_1.2.3
[169] sva_3.44.0 lattice_0.21-8 bslib_0.5.1
[172] network_1.18.1 pbkrtest_0.5.2 curl_5.0.2
[175] gtools_3.9.4 interp_1.1-4 survival_3.5-7
[178] statnet.common_4.9.0 rmarkdown_2.24 munsell_0.5.0
[181] GetoptLong_1.0.5 DO.db_2.9 GenomeInfoDbData_1.2.8
[184] iterators_1.0.14 gtable_0.3.4 rbibutils_2.2.15