Data_ind_Diffabtrial
Omar Johnson
2023-12-13
Last updated: 2024-07-23
Checks: 7 0
Knit directory: DOX_24_Github/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240723) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 85b4737. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Untracked files:
Untracked: Johnson_DOX_24_1.Rmd
Untracked: Johnson_DOX_24_2.Rmd
Untracked: Johnson_DOX_24_3.Rmd
Untracked: Johnson_DOX_24_4.Rmd
Untracked: Johnson_DOX_24_5.Rmd
Untracked: Johnson_DOX_24_7.Rmd
Untracked: Johnson_DOX_24_RUV_Limma.Rmd
Untracked: Johnson_DOX_24_RUV_Limma.html
Untracked: VennDiagram.2024-07-23_18-08-30.log
Untracked: VennDiagram.2024-07-23_18-08-31.log
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/Johnson_DOX_24_RUV_Limma.Rmd) and HTML
(docs/Johnson_DOX_24_RUV_Limma.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 85b4737 | Omar-Johnson | 2024-07-23 | more updates for paper |
| html | f37e34e | Omar-Johnson | 2024-07-23 | Build site. |
| html | e63bffa | Omar-Johnson | 2024-07-23 | Build site. |
| Rmd | 6b11493 | Omar-Johnson | 2024-07-23 | Publish Johnson_DOX_24_RUV_Limma.Rmd |
Load Libraries
Added funtions
# 1
remove_rows_with_nas <- function(data_frame) {
data_frame[rowSums(is.na(data_frame)) < 5, ]
}
# 2
remove_rows_with_nas_var <- function(data_frame, NA_remove) {
data_frame[rowSums(is.na(data_frame)) < NA_remove, ]
}
# 3
plot_nrow_vs_naremove <- function(data_frame) {
results <- data.frame(NA_remove = integer(), Rows_Remaining = integer())
for (i in 1:ncol(data_frame)) {
filtered_df <- remove_rows_with_nas_var(data_frame, NA_remove = i)
results <- rbind(results, data.frame(NA_remove = i, Rows_Remaining = nrow(filtered_df)))
}
ggplot(results, aes(x = NA_remove, y = Rows_Remaining)) +
geom_point() +
geom_line() +
xlab("Na tolerance threshold") +
ylab("Number of proteins remaining") +
theme_minimal()
}Read in Data
# Load your data
Protein_DF <- read.csv(file = "/Users/omarjohnson/Documents/Projects/Dox_Proteomics/Data/Data_Frames/RNA_Protein_DF/Data_ind_prot_DOX_24hr.csv", header = TRUE)
# Include metadata about samples
Meta <- read.csv(file = "/Users/omarjohnson/Documents/Projects/Dox_Proteomics/Data/Data_Frames/Meta_DIA.csv", header = TRUE)Wrangle Data Frame
# Save the original
Full_DF <- Protein_DF
Full_DF %>% head() Protein.Group Protein.Ids Protein.Names Genes First.Protein.Description
1 A0A0B4J2A2 A0A0B4J2A2 NA PPIAL4C NA
2 A0A0B4J2D5 A0A0B4J2D5 NA GATD3B NA
3 A0A494C071 A0A494C071 NA PWWP4 NA
4 A0AVT1 A0AVT1 NA UBA6 NA
5 A0FGR8 A0FGR8 NA ESYT2 NA
6 A0JLT2 A0JLT2 NA MED19 NA
S1 S3 S5 S7 S9 S2 S4 S6
1 979954.0 1470000 1460000 1560000 1700000 940535.0 2020000 1460000
2 16000000.0 17000000 15800000 14400000 15500000 15300000.0 13500000 13600000
3 381352.0 429735 287237 412285 380499 433456.0 540008 376304
4 62810.7 135522 159045 123045 123823 49388.1 144860 127488
5 1240000.0 1120000 969130 1040000 1020000 1120000.0 1120000 1080000
6 217977.0 181404 182657 145408 208660 224881.0 202427 160150
S8 S10
1 1390000 1620000
2 13200000 12600000
3 460580 355701
4 84684 102306
5 1120000 1060000
6 126127 209726Full_DF %>% dim()[1] 4261 15# Subset the columns referring to abundance
Protein_DF_Abundance <- Full_DF[,colnames(Full_DF) %in% c("Protein.Ids", "S1" , "S3" , "S5" , "S7", "S9", "S2", "S4", "S6" , "S8", "S10")]
Protein_DF_Abundance %>% head() Protein.Ids S1 S3 S5 S7 S9 S2
1 A0A0B4J2A2 979954.0 1470000 1460000 1560000 1700000 940535.0
2 A0A0B4J2D5 16000000.0 17000000 15800000 14400000 15500000 15300000.0
3 A0A494C071 381352.0 429735 287237 412285 380499 433456.0
4 A0AVT1 62810.7 135522 159045 123045 123823 49388.1
5 A0FGR8 1240000.0 1120000 969130 1040000 1020000 1120000.0
6 A0JLT2 217977.0 181404 182657 145408 208660 224881.0
S4 S6 S8 S10
1 2020000 1460000 1390000 1620000
2 13500000 13600000 13200000 12600000
3 540008 376304 460580 355701
4 144860 127488 84684 102306
5 1120000 1080000 1120000 1060000
6 202427 160150 126127 209726Protein_DF_Abundance %>% dim()[1] 4261 11colnames(Protein_DF_Abundance) <- c("Accession", "S1" , "S3" , "S5" , "S7", "S9", "S2", "S4", "S6" , "S8", "S10")
Protein_DF_Abundance %>% head() Accession S1 S3 S5 S7 S9 S2 S4
1 A0A0B4J2A2 979954.0 1470000 1460000 1560000 1700000 940535.0 2020000
2 A0A0B4J2D5 16000000.0 17000000 15800000 14400000 15500000 15300000.0 13500000
3 A0A494C071 381352.0 429735 287237 412285 380499 433456.0 540008
4 A0AVT1 62810.7 135522 159045 123045 123823 49388.1 144860
5 A0FGR8 1240000.0 1120000 969130 1040000 1020000 1120000.0 1120000
6 A0JLT2 217977.0 181404 182657 145408 208660 224881.0 202427
S6 S8 S10
1 1460000 1390000 1620000
2 13600000 13200000 12600000
3 376304 460580 355701
4 127488 84684 102306
5 1080000 1120000 1060000
6 160150 126127 209726Protein_DF_Abundance %>% dim()[1] 4261 11# Check for duplicated rows
# Identify duplicated values
Protein_DF_Abundance$duplicated_name <- duplicated(Protein_DF_Abundance$Accession)
# This will return a logical vector where TRUE indicates the position of duplicates in the column.
# To see only rows with duplicated values, you can subset the dataframe like this:
duplicated_rows <- Protein_DF_Abundance[Protein_DF_Abundance$duplicated_name == TRUE, ]
print(duplicated_rows) # We have 0 rows with duplicate protein names [1] Accession S1 S3 S5
[5] S7 S9 S2 S4
[9] S6 S8 S10 duplicated_name
<0 rows> (or 0-length row.names)# Make rownames of the data frame accession IDs
rownames(Protein_DF_Abundance) <- Protein_DF_Abundance$Accession
# Change colnames
colnames(Protein_DF_Abundance) [1] "Accession" "S1" "S3" "S5"
[5] "S7" "S9" "S2" "S4"
[9] "S6" "S8" "S10" "duplicated_name"Protein_DF_Abundance <- Protein_DF_Abundance[, -c(1, 12)]
Protein_DF_Abundance %>% head() S1 S3 S5 S7 S9 S2 S4
A0A0B4J2A2 979954.0 1470000 1460000 1560000 1700000 940535.0 2020000
A0A0B4J2D5 16000000.0 17000000 15800000 14400000 15500000 15300000.0 13500000
A0A494C071 381352.0 429735 287237 412285 380499 433456.0 540008
A0AVT1 62810.7 135522 159045 123045 123823 49388.1 144860
A0FGR8 1240000.0 1120000 969130 1040000 1020000 1120000.0 1120000
A0JLT2 217977.0 181404 182657 145408 208660 224881.0 202427
S6 S8 S10
A0A0B4J2A2 1460000 1390000 1620000
A0A0B4J2D5 13600000 13200000 12600000
A0A494C071 376304 460580 355701
A0AVT1 127488 84684 102306
A0FGR8 1080000 1120000 1060000
A0JLT2 160150 126127 209726Protein_DF_Abundance %>% dim()[1] 4261 10# Assuming column names of Protein_DF match with the rows in Meta
Meta Samples Ind Rep Cond Cond_Ind
1 S1 B_77 Bio Dox B_77_Dox
2 S3 C_87 Bio Dox C_87_Dox
3 S5 A_48 Tech Dox A_48_Dox
4 S7 A_48 Tech Dox A_48_Dox
5 S9 A_48 Tech Dox A_48_Dox
6 S2 B_77 Bio Control B_77_Control
7 S4 C_87 Bio Control C_87_Control
8 S6 A_48 Tech Control A_48_Control
9 S8 A_48 Tech Control A_48_Control
10 S10 A_48 Tech Control A_48_Controlrownames(Meta) <- Meta$Samples
Meta Samples Ind Rep Cond Cond_Ind
S1 S1 B_77 Bio Dox B_77_Dox
S3 S3 C_87 Bio Dox C_87_Dox
S5 S5 A_48 Tech Dox A_48_Dox
S7 S7 A_48 Tech Dox A_48_Dox
S9 S9 A_48 Tech Dox A_48_Dox
S2 S2 B_77 Bio Control B_77_Control
S4 S4 C_87 Bio Control C_87_Control
S6 S6 A_48 Tech Control A_48_Control
S8 S8 A_48 Tech Control A_48_Control
S10 S10 A_48 Tech Control A_48_Controlcolnames(Protein_DF_Abundance) <- Meta$Samples
Protein_DF_Abundance %>% head() S1 S3 S5 S7 S9 S2 S4
A0A0B4J2A2 979954.0 1470000 1460000 1560000 1700000 940535.0 2020000
A0A0B4J2D5 16000000.0 17000000 15800000 14400000 15500000 15300000.0 13500000
A0A494C071 381352.0 429735 287237 412285 380499 433456.0 540008
A0AVT1 62810.7 135522 159045 123045 123823 49388.1 144860
A0FGR8 1240000.0 1120000 969130 1040000 1020000 1120000.0 1120000
A0JLT2 217977.0 181404 182657 145408 208660 224881.0 202427
S6 S8 S10
A0A0B4J2A2 1460000 1390000 1620000
A0A0B4J2D5 13600000 13200000 12600000
A0A494C071 376304 460580 355701
A0AVT1 127488 84684 102306
A0FGR8 1080000 1120000 1060000
A0JLT2 160150 126127 209726Meta Samples Ind Rep Cond Cond_Ind
S1 S1 B_77 Bio Dox B_77_Dox
S3 S3 C_87 Bio Dox C_87_Dox
S5 S5 A_48 Tech Dox A_48_Dox
S7 S7 A_48 Tech Dox A_48_Dox
S9 S9 A_48 Tech Dox A_48_Dox
S2 S2 B_77 Bio Control B_77_Control
S4 S4 C_87 Bio Control C_87_Control
S6 S6 A_48 Tech Control A_48_Control
S8 S8 A_48 Tech Control A_48_Control
S10 S10 A_48 Tech Control A_48_ControlRemove NA
#1. Find rows with any NA values
rows_with_na <- apply(Protein_DF_Abundance, 1, function(x) any(is.na(x))) %>% which()
rows_with_na <- as.numeric(rows_with_na)
# 2. Remove rows with NA values
Protein_DF_Abundance <- Protein_DF_Abundance[-rows_with_na, ]
# 3. Sanity check
Protein_DF_Abundance %>% dim()[1] 3934 10Log2 transform
log2_norm_counts <- log2(Protein_DF_Abundance)
log2_norm_counts %>% rowMeans() %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Quantile Normalization
normalizedData <- limma::normalizeBetweenArrays(log2_norm_counts[, c(1,2,4,6,7,9)], method = "quantile")
normalizedData %>% rowMeans() %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Linear model - RUVs
# counts need to be integer values and in a numeric matrix
counts <- as.matrix(normalizedData)
counts %>% dim()[1] 3934 6# Create a DataFrame for the phenoData
phenoData <- DataFrame(Meta)
phenoData_sub <- phenoData[c(1,2,4,6,7,9), ]
# Create Design Matrix
# phenoData$Cond <- factor(phenoData$Cond , levels = c("Control", "Dox"))
design <- model.matrix(~ 0 + Cond , phenoData_sub)
design CondControl CondDox
S1 0 1
S3 0 1
S7 0 1
S2 1 0
S4 1 0
S8 1 0
attr(,"assign")
[1] 1 1
attr(,"contrasts")
attr(,"contrasts")$Cond
[1] "contr.treatment"# rename columns
colnames(design) <- c('Control', "Dox")
# Fit model
dupcor <- duplicateCorrelation(counts , design, block = phenoData_sub$Ind)
fit <- lmFit(object = counts, design = design, block = phenoData_sub$Ind, correlation = dupcor$consensus)
fit2 <- eBayes(fit)
# Make contrasts
cm <- makeContrasts(
DoxvNorm = Dox - Control,
levels = design)
# Model with contrasts
fit2 <- contrasts.fit(fit, cm)
fit2 <- eBayes(fit2 )
# Summarize
results_summary <- decideTests(fit2, adjust.method = "none", p.value = 0.05)
summary(results_summary) DoxvNorm
Down 570
NotSig 2835
Up 529# Toptable summary organized to contain results for all tested proteins
toptable_summary <- topTable(fit2, coef = "DoxvNorm",number = (nrow(normalizedData)), p.value = 1, adjust.method = "none")
toptable_summary$Protein <- rownames(toptable_summary)
toptable_summary$P.Value %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# PCA of log2-quantile normalized-RUVs values:
prcomp_res <- prcomp(t(normalizedData %>% as.matrix()), center = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(phenoData_sub), colour = "Cond", shape = "Ind", size =4)+
theme_bw()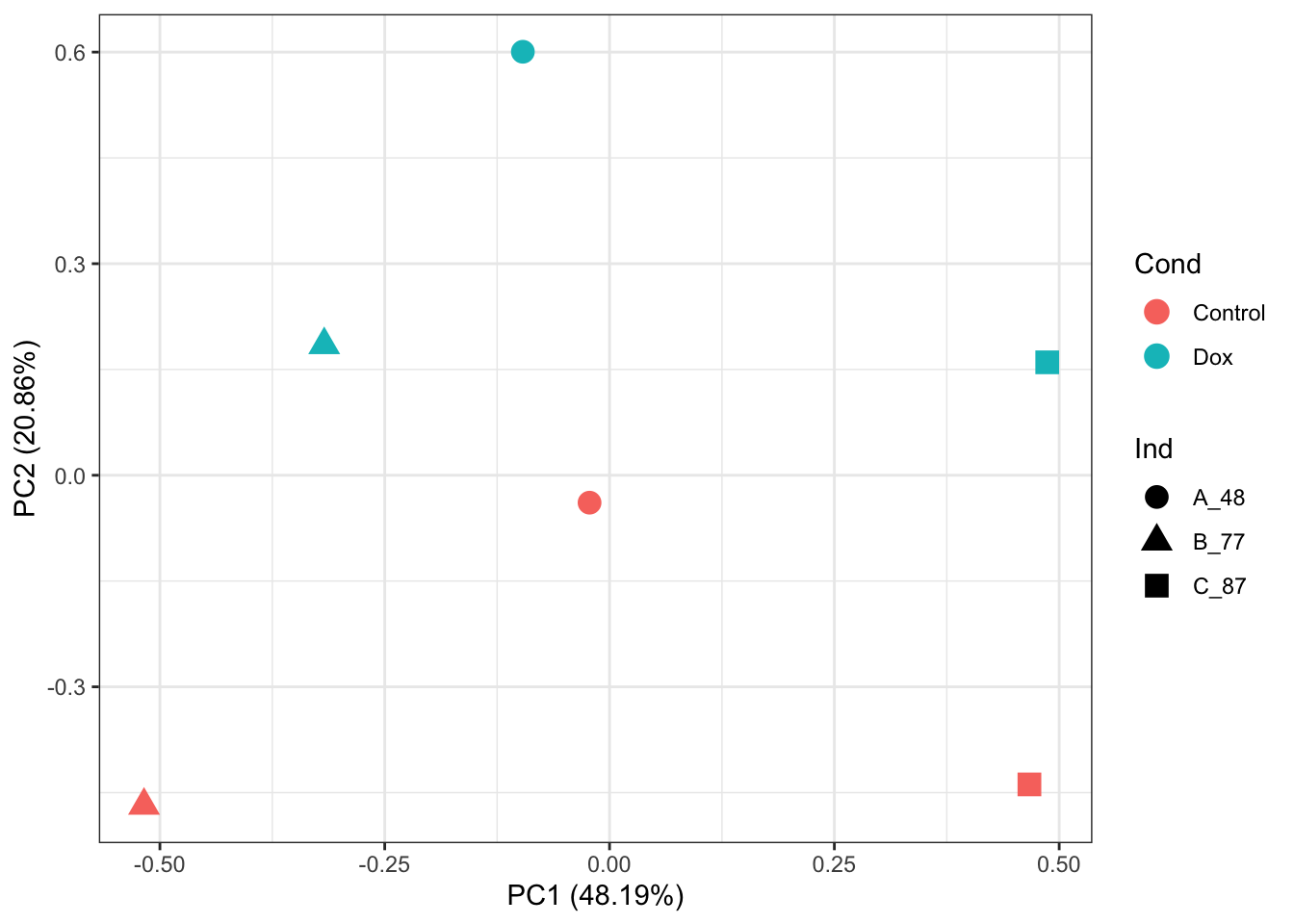
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Volcano plots
# 1. Create a column to threshold P-values
toptable_summary <- toptable_summary %>% mutate(threshold_P = P.Value < 0.05)
# 2. Plot
ggplot(toptable_summary)+
geom_point(mapping = aes(x = logFC, y = -log10(P.Value), color = threshold_P))+
xlab("log2FC")+
ylab("-log10 nominal p-value")+
ylim(0, 7.5)+
xlim(-5, 5)+
theme(legend.position = "none",
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = rel(1.25)))+
theme_bw()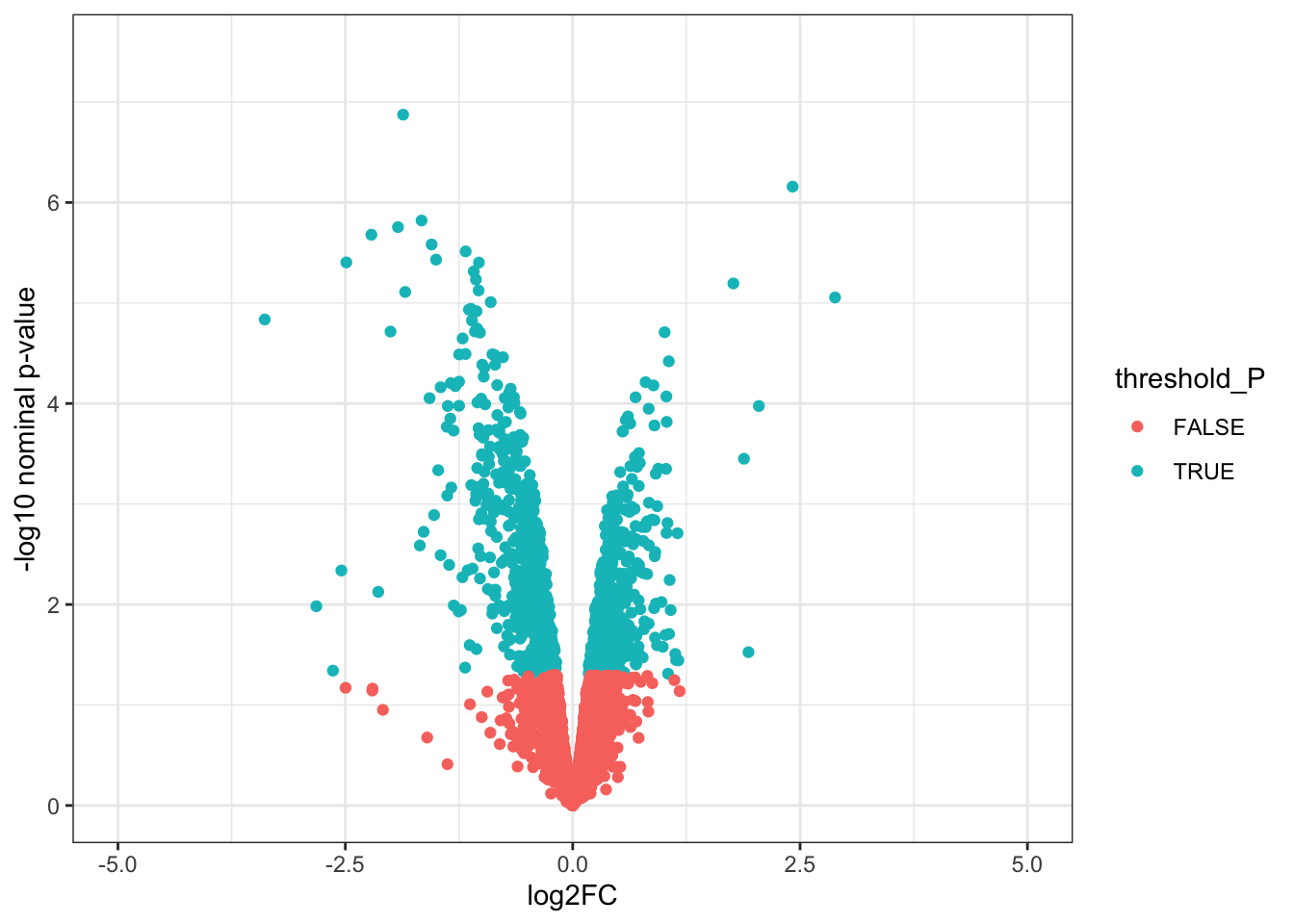
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Other fits
fit3 <- eBayes(fit2, robust = TRUE, trend = TRUE)Warning: One very small variance detected, has been offset away from zero# Summarize
results_summary <- decideTests(fit3, adjust.method = "none", p.value = 0.05)
summary(results_summary) DoxvNorm
Down 572
NotSig 2808
Up 554# Toptable summary organized to contain results for all tested proteins
toptable_summary <- topTable(fit3, coef = "DoxvNorm",number = (nrow(normalizedData)), p.value = 1, adjust.method = "none")
toptable_summary$Protein <- rownames(toptable_summary)
toptable_summary$P.Value %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# PCA of log2-quantile normalized-RUVs values:
prcomp_res <- prcomp(t(normalizedData %>% as.matrix()), center = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(phenoData_sub), colour = "Cond", shape = "Ind", size =4)+
theme_bw()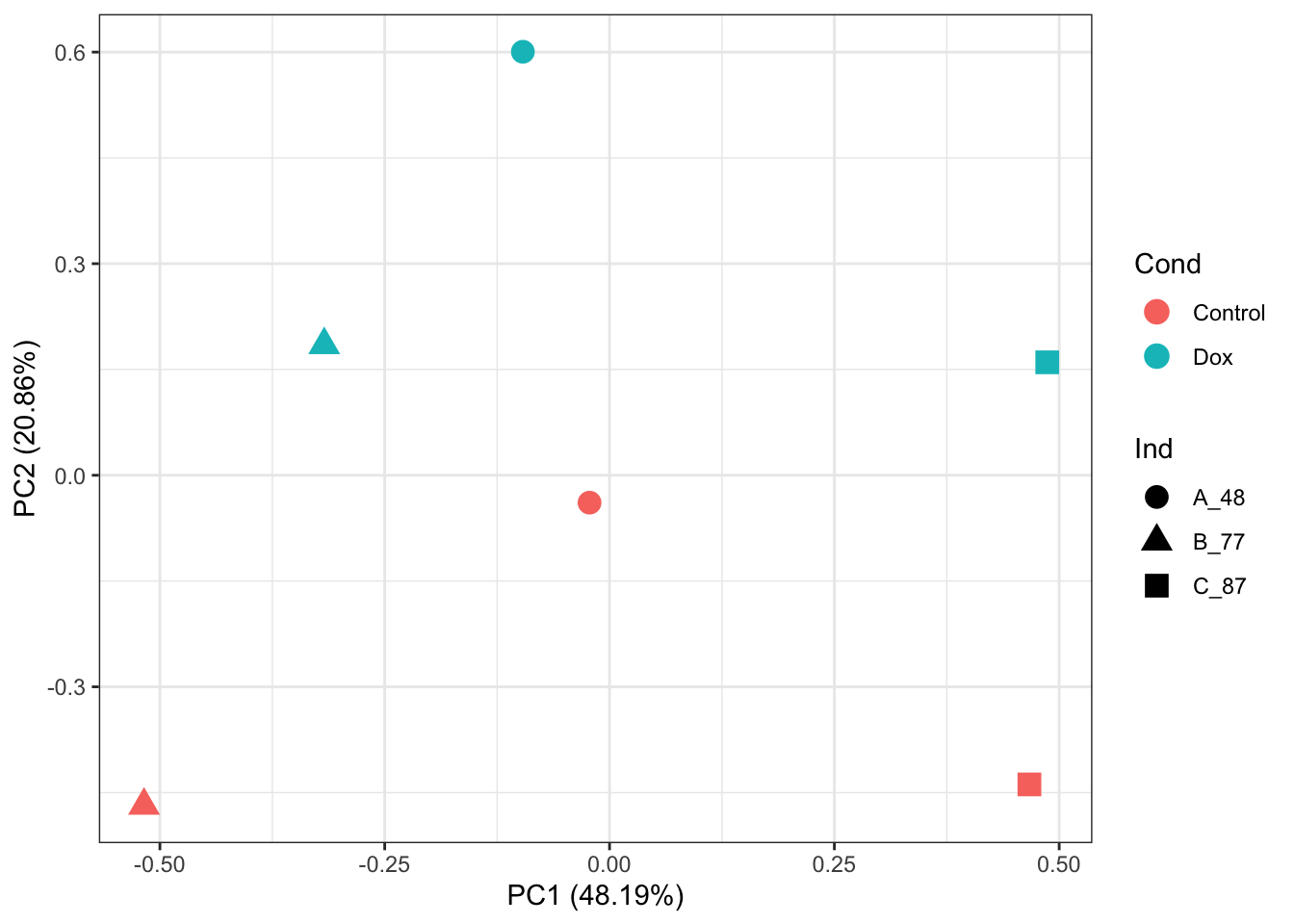
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Volcano plots
# 1. Create a column to threshold P-values
toptable_summary <- toptable_summary %>% mutate(threshold_P = P.Value < 0.05)
# 2. Plot
ggplot(toptable_summary)+
geom_point(mapping = aes(x = logFC, y = -log10(P.Value), color = threshold_P))+
xlab("log2FC")+
ylab("-log10 nominal p-value")+
ylim(0, 7.5)+
xlim(-5, 5)+
theme(legend.position = "none",
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = rel(1.25)))+
theme_bw()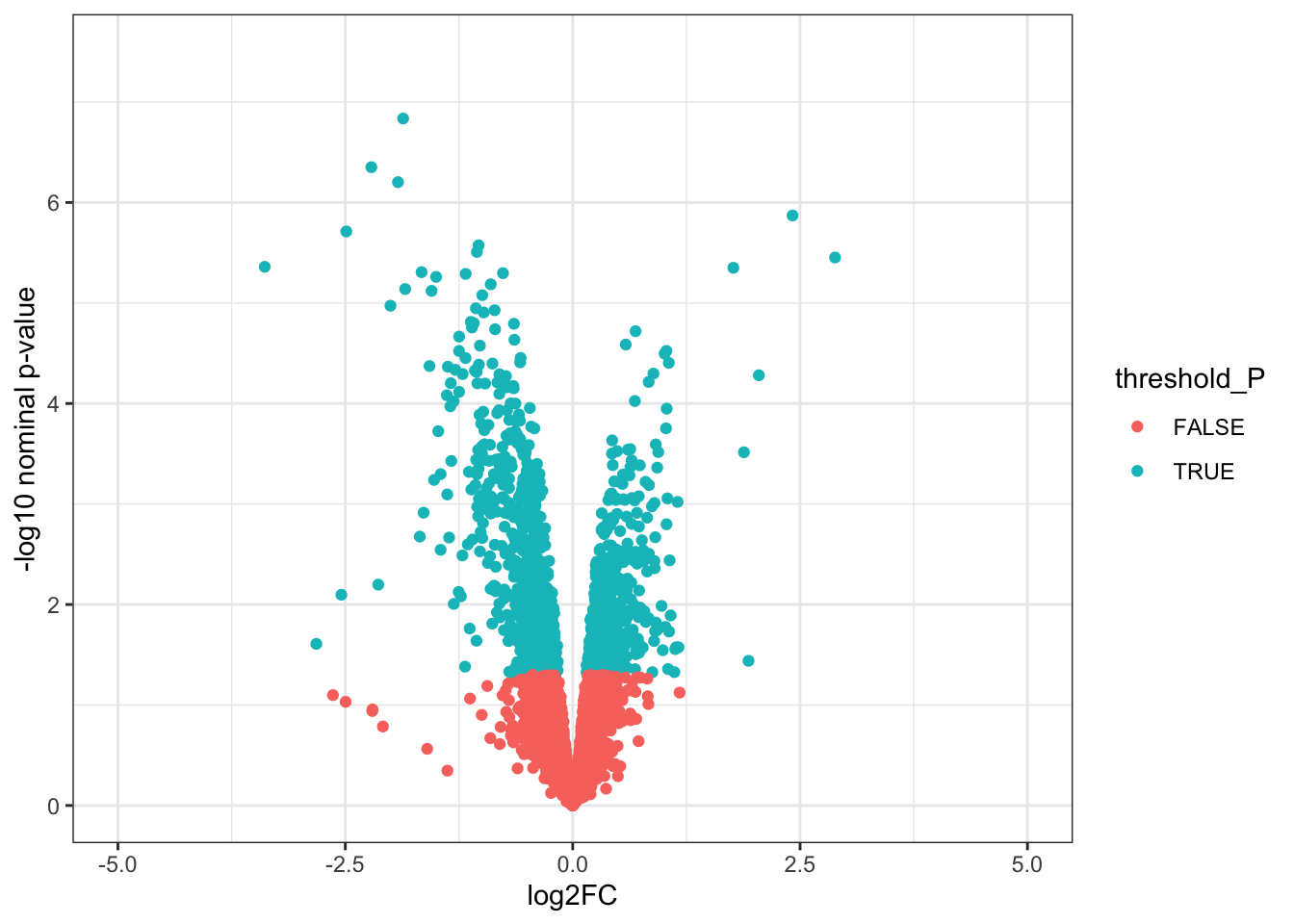
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Wrangle Data Frame for imputed data
# Save the original
Full_DF <- Protein_DF
Full_DF %>% head() Protein.Group Protein.Ids Protein.Names Genes First.Protein.Description
1 A0A0B4J2A2 A0A0B4J2A2 NA PPIAL4C NA
2 A0A0B4J2D5 A0A0B4J2D5 NA GATD3B NA
3 A0A494C071 A0A494C071 NA PWWP4 NA
4 A0AVT1 A0AVT1 NA UBA6 NA
5 A0FGR8 A0FGR8 NA ESYT2 NA
6 A0JLT2 A0JLT2 NA MED19 NA
S1 S3 S5 S7 S9 S2 S4 S6
1 979954.0 1470000 1460000 1560000 1700000 940535.0 2020000 1460000
2 16000000.0 17000000 15800000 14400000 15500000 15300000.0 13500000 13600000
3 381352.0 429735 287237 412285 380499 433456.0 540008 376304
4 62810.7 135522 159045 123045 123823 49388.1 144860 127488
5 1240000.0 1120000 969130 1040000 1020000 1120000.0 1120000 1080000
6 217977.0 181404 182657 145408 208660 224881.0 202427 160150
S8 S10
1 1390000 1620000
2 13200000 12600000
3 460580 355701
4 84684 102306
5 1120000 1060000
6 126127 209726Full_DF %>% dim()[1] 4261 15# # Subset the columns referring to abundance
Protein_DF_Abundance <- Full_DF[,colnames(Full_DF) %in% c("Protein.Ids", "S1" , "S3" , "S5" , "S7", "S9", "S2", "S4", "S6" , "S8", "S10")]
Protein_DF_Abundance %>% head() Protein.Ids S1 S3 S5 S7 S9 S2
1 A0A0B4J2A2 979954.0 1470000 1460000 1560000 1700000 940535.0
2 A0A0B4J2D5 16000000.0 17000000 15800000 14400000 15500000 15300000.0
3 A0A494C071 381352.0 429735 287237 412285 380499 433456.0
4 A0AVT1 62810.7 135522 159045 123045 123823 49388.1
5 A0FGR8 1240000.0 1120000 969130 1040000 1020000 1120000.0
6 A0JLT2 217977.0 181404 182657 145408 208660 224881.0
S4 S6 S8 S10
1 2020000 1460000 1390000 1620000
2 13500000 13600000 13200000 12600000
3 540008 376304 460580 355701
4 144860 127488 84684 102306
5 1120000 1080000 1120000 1060000
6 202427 160150 126127 209726Protein_DF_Abundance %>% dim()[1] 4261 11colnames(Protein_DF_Abundance) <- c("Accession", "S1" , "S3" , "S5" , "S7", "S9", "S2", "S4", "S6" , "S8", "S10")
Protein_DF_Abundance %>% head() Accession S1 S3 S5 S7 S9 S2 S4
1 A0A0B4J2A2 979954.0 1470000 1460000 1560000 1700000 940535.0 2020000
2 A0A0B4J2D5 16000000.0 17000000 15800000 14400000 15500000 15300000.0 13500000
3 A0A494C071 381352.0 429735 287237 412285 380499 433456.0 540008
4 A0AVT1 62810.7 135522 159045 123045 123823 49388.1 144860
5 A0FGR8 1240000.0 1120000 969130 1040000 1020000 1120000.0 1120000
6 A0JLT2 217977.0 181404 182657 145408 208660 224881.0 202427
S6 S8 S10
1 1460000 1390000 1620000
2 13600000 13200000 12600000
3 376304 460580 355701
4 127488 84684 102306
5 1080000 1120000 1060000
6 160150 126127 209726Protein_DF_Abundance %>% dim()[1] 4261 11# # Check for duplicated rows
# # Identify duplicated values
Protein_DF_Abundance$duplicated_name <- duplicated(Protein_DF_Abundance$Accession)
# # This will return a logical vector where TRUE indicates the position of duplicates in the column.
# # To see only rows with duplicated values, you can subset the dataframe like this:
duplicated_rows <- Protein_DF_Abundance[Protein_DF_Abundance$duplicated_name == TRUE, ]
print(duplicated_rows) # We have 0 rows with duplicate protein names [1] Accession S1 S3 S5
[5] S7 S9 S2 S4
[9] S6 S8 S10 duplicated_name
<0 rows> (or 0-length row.names)# # Make rownames of the data frame accession IDs
rownames(Protein_DF_Abundance) <- Protein_DF_Abundance$Accession
# # Change colnames
colnames(Protein_DF_Abundance) [1] "Accession" "S1" "S3" "S5"
[5] "S7" "S9" "S2" "S4"
[9] "S6" "S8" "S10" "duplicated_name"Protein_DF_Abundance <- Protein_DF_Abundance[, -c(1, 12)]
Protein_DF_Abundance %>% head() S1 S3 S5 S7 S9 S2 S4
A0A0B4J2A2 979954.0 1470000 1460000 1560000 1700000 940535.0 2020000
A0A0B4J2D5 16000000.0 17000000 15800000 14400000 15500000 15300000.0 13500000
A0A494C071 381352.0 429735 287237 412285 380499 433456.0 540008
A0AVT1 62810.7 135522 159045 123045 123823 49388.1 144860
A0FGR8 1240000.0 1120000 969130 1040000 1020000 1120000.0 1120000
A0JLT2 217977.0 181404 182657 145408 208660 224881.0 202427
S6 S8 S10
A0A0B4J2A2 1460000 1390000 1620000
A0A0B4J2D5 13600000 13200000 12600000
A0A494C071 376304 460580 355701
A0AVT1 127488 84684 102306
A0FGR8 1080000 1120000 1060000
A0JLT2 160150 126127 209726Protein_DF_Abundance %>% dim()[1] 4261 10#
# # Assuming column names of Protein_DF match with the rows in Meta
Meta Samples Ind Rep Cond Cond_Ind
S1 S1 B_77 Bio Dox B_77_Dox
S3 S3 C_87 Bio Dox C_87_Dox
S5 S5 A_48 Tech Dox A_48_Dox
S7 S7 A_48 Tech Dox A_48_Dox
S9 S9 A_48 Tech Dox A_48_Dox
S2 S2 B_77 Bio Control B_77_Control
S4 S4 C_87 Bio Control C_87_Control
S6 S6 A_48 Tech Control A_48_Control
S8 S8 A_48 Tech Control A_48_Control
S10 S10 A_48 Tech Control A_48_Controlrownames(Meta) <- Meta$Samples
Meta Samples Ind Rep Cond Cond_Ind
S1 S1 B_77 Bio Dox B_77_Dox
S3 S3 C_87 Bio Dox C_87_Dox
S5 S5 A_48 Tech Dox A_48_Dox
S7 S7 A_48 Tech Dox A_48_Dox
S9 S9 A_48 Tech Dox A_48_Dox
S2 S2 B_77 Bio Control B_77_Control
S4 S4 C_87 Bio Control C_87_Control
S6 S6 A_48 Tech Control A_48_Control
S8 S8 A_48 Tech Control A_48_Control
S10 S10 A_48 Tech Control A_48_Controlcolnames(Protein_DF_Abundance) <- Meta$Samples
Protein_DF_Abundance %>% head() S1 S3 S5 S7 S9 S2 S4
A0A0B4J2A2 979954.0 1470000 1460000 1560000 1700000 940535.0 2020000
A0A0B4J2D5 16000000.0 17000000 15800000 14400000 15500000 15300000.0 13500000
A0A494C071 381352.0 429735 287237 412285 380499 433456.0 540008
A0AVT1 62810.7 135522 159045 123045 123823 49388.1 144860
A0FGR8 1240000.0 1120000 969130 1040000 1020000 1120000.0 1120000
A0JLT2 217977.0 181404 182657 145408 208660 224881.0 202427
S6 S8 S10
A0A0B4J2A2 1460000 1390000 1620000
A0A0B4J2D5 13600000 13200000 12600000
A0A494C071 376304 460580 355701
A0AVT1 127488 84684 102306
A0FGR8 1080000 1120000 1060000
A0JLT2 160150 126127 209726Meta Samples Ind Rep Cond Cond_Ind
S1 S1 B_77 Bio Dox B_77_Dox
S3 S3 C_87 Bio Dox C_87_Dox
S5 S5 A_48 Tech Dox A_48_Dox
S7 S7 A_48 Tech Dox A_48_Dox
S9 S9 A_48 Tech Dox A_48_Dox
S2 S2 B_77 Bio Control B_77_Control
S4 S4 C_87 Bio Control C_87_Control
S6 S6 A_48 Tech Control A_48_Control
S8 S8 A_48 Tech Control A_48_Control
S10 S10 A_48 Tech Control A_48_ControlFigS2A Viewing how NA threshold vs remaining proteins
# View how NA thresholds affect the amount of proteins remaining to model
# All proteins
Protein_DF_Abundance %>% nrow()[1] 4261# Removing proteins with 1 or more NA
remove_rows_with_nas_var(data_frame = Protein_DF_Abundance , NA_remove = 1 ) %>% nrow()[1] 3934# Removing proteins with 2 or more NA
remove_rows_with_nas_var(data_frame = Protein_DF_Abundance , NA_remove = 2 ) %>% nrow()[1] 4051# Removing proteins with 3 or more NA
remove_rows_with_nas_var(data_frame = Protein_DF_Abundance , NA_remove = 3 ) %>% nrow()[1] 4118# Removing proteins with 4 or more NA
remove_rows_with_nas_var(data_frame = Protein_DF_Abundance , NA_remove = 4 ) %>% nrow()[1] 4153# Removing proteins with 5 or more NA
remove_rows_with_nas_var(data_frame = Protein_DF_Abundance , NA_remove = 5 ) %>% nrow()[1] 4182# Removing proteins with 6 or more NA
remove_rows_with_nas_var(data_frame = Protein_DF_Abundance , NA_remove = 6 ) %>% nrow()[1] 4212# Removing proteins with 7 or more NA
remove_rows_with_nas_var(data_frame = Protein_DF_Abundance , NA_remove = 7 ) %>% nrow()[1] 4229# Removing proteins with 8 or more NA
remove_rows_with_nas_var(data_frame = Protein_DF_Abundance , NA_remove = 8 ) %>% nrow()[1] 4241# View in graph
plot_nrow_vs_naremove(Protein_DF_Abundance)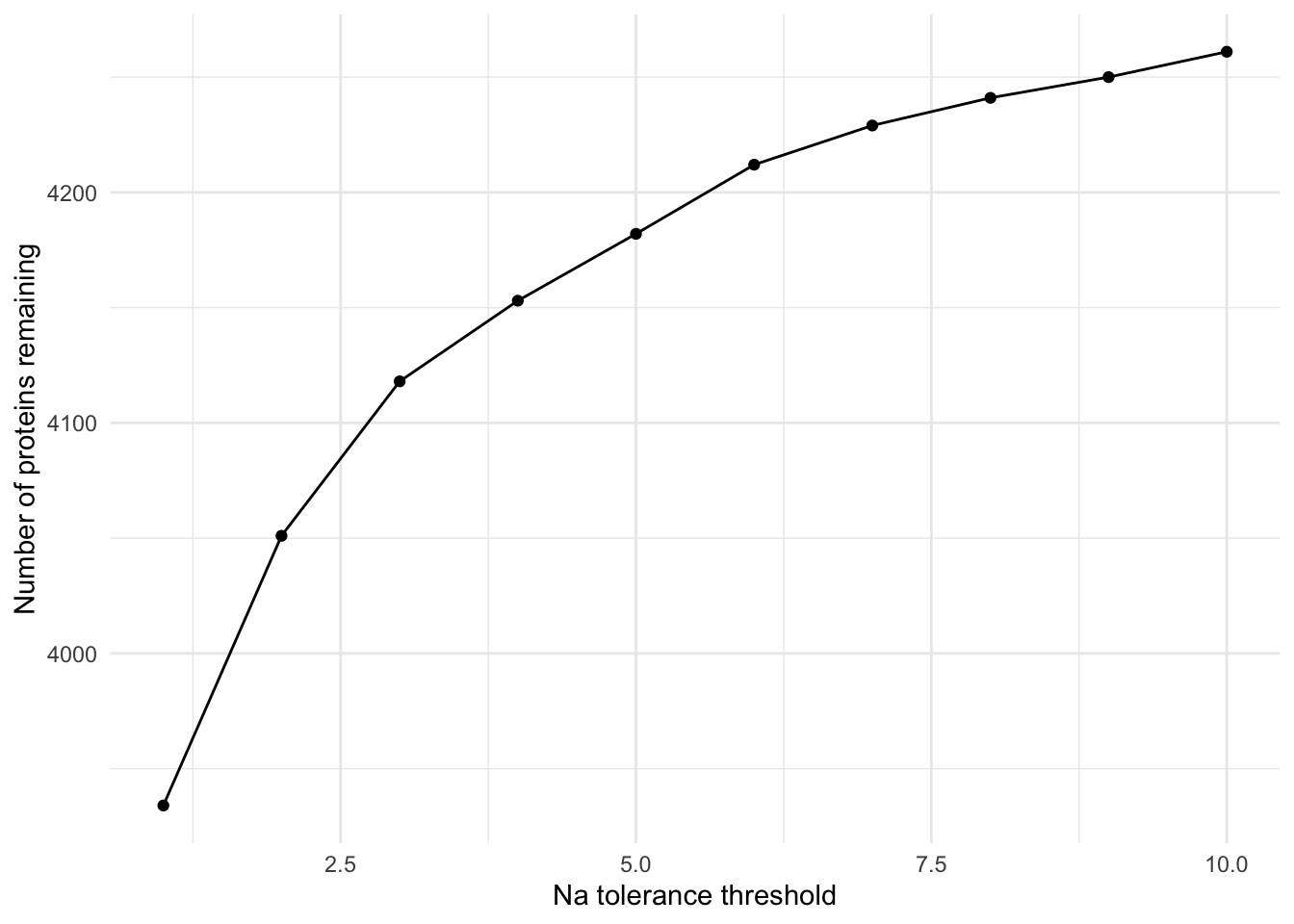
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
NA removal and imputing with knn
# View original abundances
Protein_DF_Abundance %>% head() S1 S3 S5 S7 S9 S2 S4
A0A0B4J2A2 979954.0 1470000 1460000 1560000 1700000 940535.0 2020000
A0A0B4J2D5 16000000.0 17000000 15800000 14400000 15500000 15300000.0 13500000
A0A494C071 381352.0 429735 287237 412285 380499 433456.0 540008
A0AVT1 62810.7 135522 159045 123045 123823 49388.1 144860
A0FGR8 1240000.0 1120000 969130 1040000 1020000 1120000.0 1120000
A0JLT2 217977.0 181404 182657 145408 208660 224881.0 202427
S6 S8 S10
A0A0B4J2A2 1460000 1390000 1620000
A0A0B4J2D5 13600000 13200000 12600000
A0A494C071 376304 460580 355701
A0AVT1 127488 84684 102306
A0FGR8 1080000 1120000 1060000
A0JLT2 160150 126127 209726Protein_DF_Abundance[,1] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance[,3] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance[,4] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance[,5] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance[,6] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance[,7] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance[,8] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance[,9] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance[,10] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# View abundances after removing proteins with 5 or more (50%) NA
Protein_DF_Abundance_imp <- remove_rows_with_nas(Protein_DF_Abundance)
Protein_DF_Abundance_imp %>% dim()[1] 4182 10Protein_DF_Abundance_imp[,1] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp[,3] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp[,4] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp[,5] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp[,6] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp[,7] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp[,8] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp[,9] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp[,10] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp_2 <- impute.knn(as.matrix(Protein_DF_Abundance_imp), k = 10, rowmax = 0.4)$dataCluster size 4182 broken into 60 4122
Done cluster 60
Cluster size 4122 broken into 316 3806
Done cluster 316
Cluster size 3806 broken into 2988 818
Cluster size 2988 broken into 2086 902
Cluster size 2086 broken into 843 1243
Done cluster 843
Done cluster 1243
Done cluster 2086
Done cluster 902
Done cluster 2988
Done cluster 818
Done cluster 3806
Done cluster 4122 Protein_DF_Abundance_imp_2 %>% dim()[1] 4182 10Protein_DF_Abundance_imp_2[,1] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp_2[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp_2[,3] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp_2[,4] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp_2[,5] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp_2[,6] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp_2[,7] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp_2[,8] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp_2[,9] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Protein_DF_Abundance_imp_2[,10] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Log2 transform for imputed data
log2_norm_counts <- log2(Protein_DF_Abundance_imp_2)
log2_norm_counts %>% rowMeans() %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
log2_norm_counts[,1] %>% hist(breaks = 100) 
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
log2_norm_counts[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
log2_norm_counts[,3] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
log2_norm_counts[,4] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
log2_norm_counts[,5] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
log2_norm_counts[,6] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
log2_norm_counts[,7] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
log2_norm_counts[,8] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
log2_norm_counts[,9] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
log2_norm_counts[,10] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Quantile Normalization for imputed data
normalizedData <- limma::normalizeBetweenArrays(log2_norm_counts[, c(1,2,4,6,7,9)], method = "quantile")
normalizedData %>% rowMeans() %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
normalizedData[,1] %>% hist(breaks = 100) 
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
normalizedData[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
normalizedData[,3] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
normalizedData[,4] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
normalizedData[,5] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
normalizedData[,6] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
FigS2 B View distribution of imputed NA values for all samples
na_positions <- is.na(Protein_DF_Abundance_imp)
imputed_dataframe <- normalizedData
imputed_values <- imputed_dataframe[na_positions] %>% na.omit()
hist(imputed_values, breaks = 50, main = "Histogram of Imputed Values", xlab = "Value")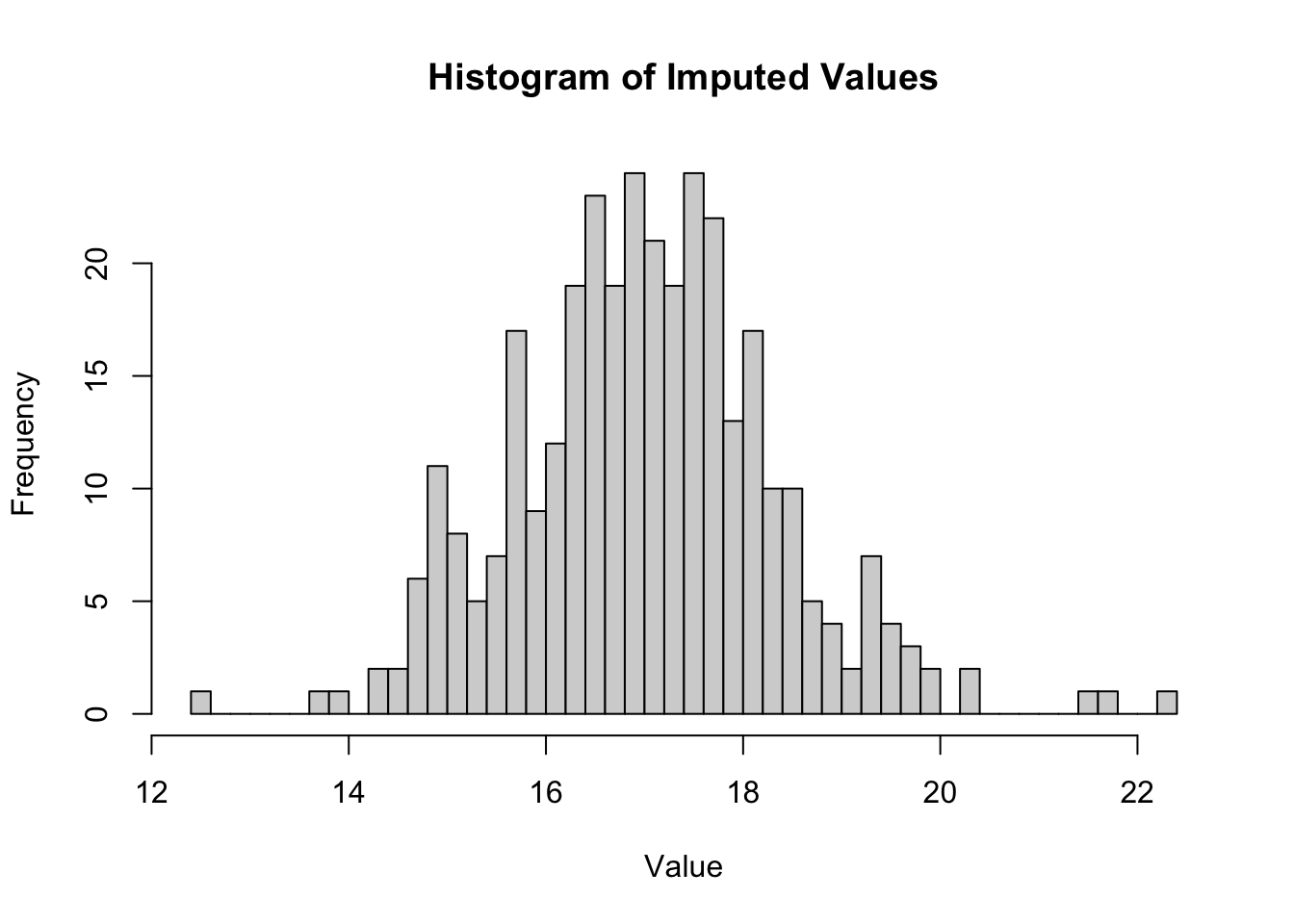
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Histogram of original data (with NA values removed)
hist(log2(Protein_DF_Abundance_imp) %>% rowMeans(na.rm = TRUE), xlim = range(log2(Protein_DF_Abundance_imp) %>% rowMeans(), na.rm = TRUE), breaks = 50, main = "Histogram with Imputed & Non-imputed Values", xlab = "Value")
# Histogram of imputed values
hist(imputed_values, breaks = 50, add = TRUE, col = "red")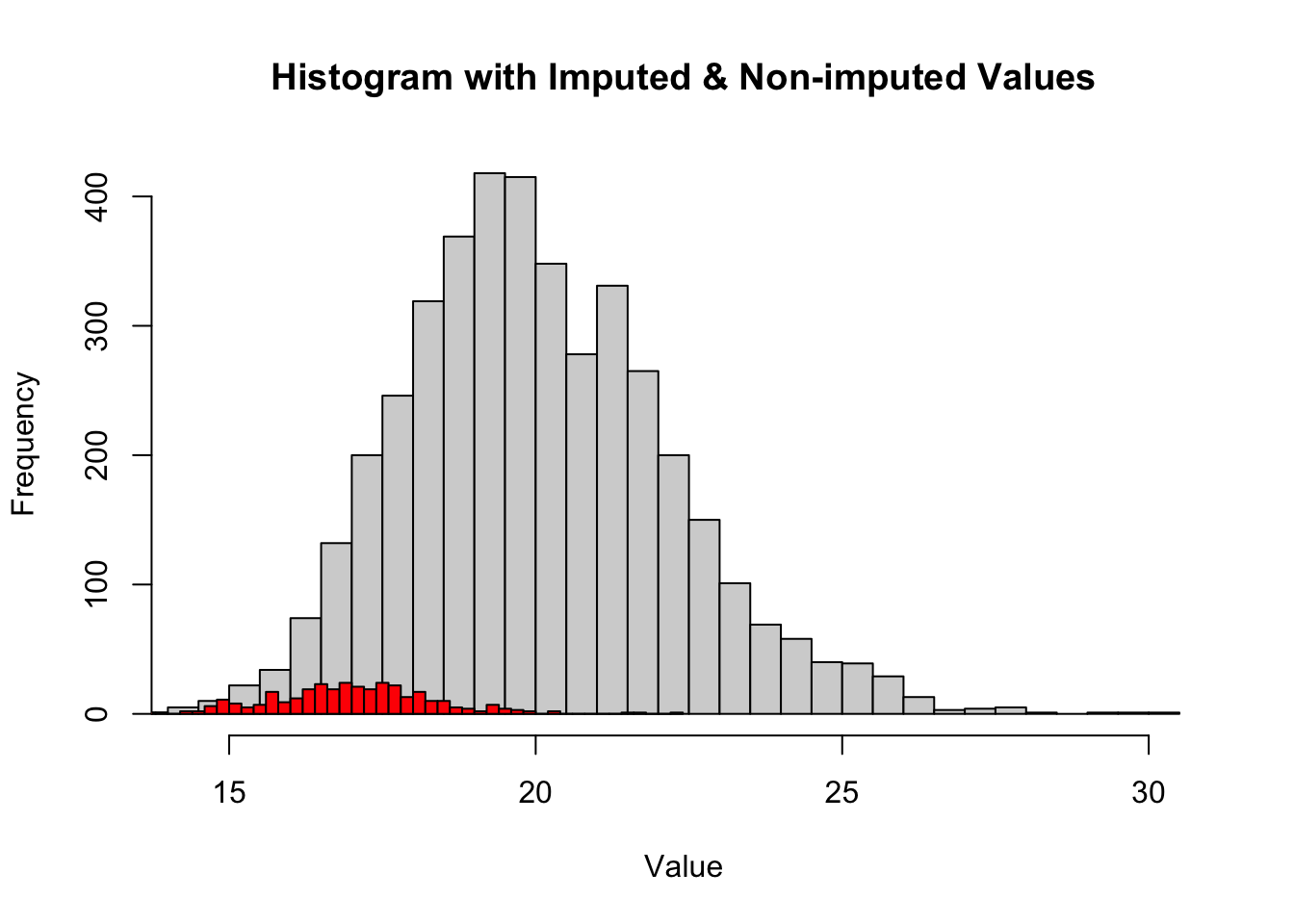
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Linear model - RUVs + Imputation
# counts need to be integer values and in a numeric matrix
counts <- as.matrix(normalizedData)
counts %>% dim()[1] 4182 6# Create a DataFrame for the phenoData
phenoData <- DataFrame(Meta)
phenoData_sub <- phenoData[c(1,2,4,6,7,9), ]
# Create Design Matrix
# phenoData$Cond <- factor(phenoData$Cond , levels = c("Control", "Dox"))
design <- model.matrix(~ 0 + Cond , phenoData_sub)
design CondControl CondDox
S1 0 1
S3 0 1
S7 0 1
S2 1 0
S4 1 0
S8 1 0
attr(,"assign")
[1] 1 1
attr(,"contrasts")
attr(,"contrasts")$Cond
[1] "contr.treatment"# rename columns
colnames(design) <- c('Control', "Dox")
# Fit model
dupcor <- duplicateCorrelation(counts , design, block = phenoData_sub$Ind)
fit <- lmFit(object = counts, design = design, block = phenoData_sub$Ind, correlation = dupcor$consensus)
fit2 <- eBayes(fit)
# Make contrasts
cm <- makeContrasts(
DoxvNorm = Dox - Control,
levels = design)
# Model with contrasts
fit2 <- contrasts.fit(fit, cm)
fit2 <- eBayes(fit2 )
# Summarize
results_summary <- decideTests(fit2, adjust.method = "none", p.value = 0.05)
summary(results_summary) DoxvNorm
Down 578
NotSig 3061
Up 543# Toptable summary organized to contain results for all tested proteins
toptable_summary <- topTable(fit2, coef = "DoxvNorm",number = (nrow(normalizedData)), p.value = 1, adjust.method = "none")
toptable_summary$Protein <- rownames(toptable_summary)
toptable_summary$P.Value %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# PCA of log2-quantile normalized-RUVs values:
prcomp_res <- prcomp(t(normalizedData %>% as.matrix()), center = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(phenoData_sub), colour = "Cond", shape = "Ind", size =4)+
theme_bw()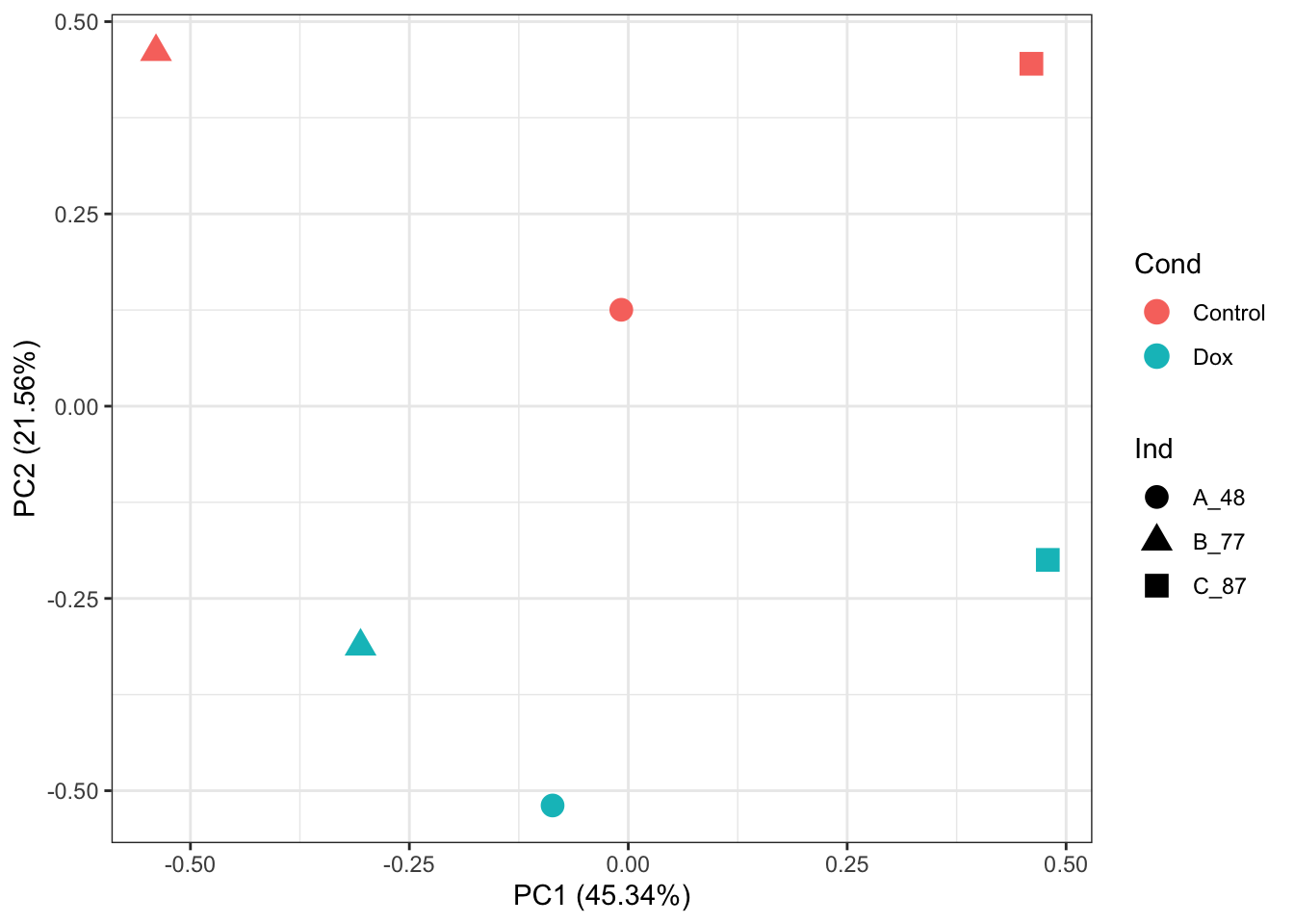
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Volcano plots
# 1. Create a column to threshold P-values
toptable_summary <- toptable_summary %>% mutate(threshold_P = P.Value < 0.05)
# 2. Plot
ggplot(toptable_summary)+
geom_point(mapping = aes(x = logFC, y = -log10(P.Value), color = threshold_P))+
xlab("log2FC")+
ylab("-log10 nominal p-value")+
ylim(0, 10)+
xlim(-7, 7)+
theme(legend.position = "none",
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = rel(1.25)))+
theme_bw()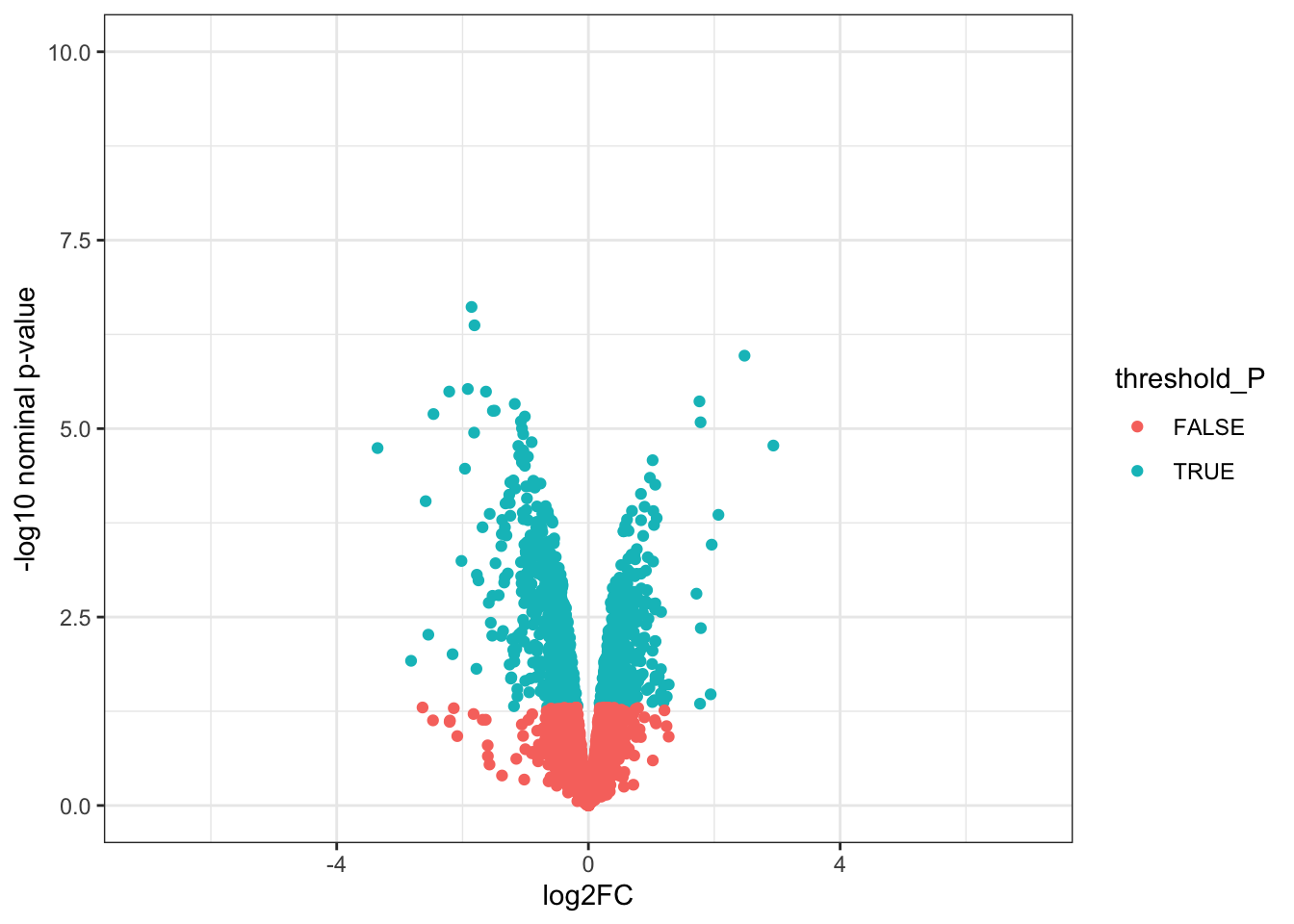
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Other fits
fit3 <- eBayes(fit2, robust = TRUE, trend = TRUE)Warning: One very small variance detected, has been offset away from zero# Summarize
results_summary <- decideTests(fit3, adjust.method = "none", p.value = 0.05)
summary(results_summary) DoxvNorm
Down 584
NotSig 3033
Up 565# Toptable summary organized to contain results for all tested proteins
toptable_summary <- topTable(fit3, coef = "DoxvNorm",number = (nrow(normalizedData)), p.value = 1, adjust.method = "none")
toptable_summary$Protein <- rownames(toptable_summary)
toptable_summary$P.Value %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
toptable_summary["P16860",] logFC AveExpr t P.Value adj.P.Val B Protein
P16860 -0.0824422 17.29226 -0.1403269 0.8922319 0.8922319 -6.866182 P16860# PCA of log2-quantile normalized-RUVs values:
prcomp_res <- prcomp(t(normalizedData %>% as.matrix()), center = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(phenoData_sub), colour = "Cond", shape = "Ind", size =4)+
theme_bw()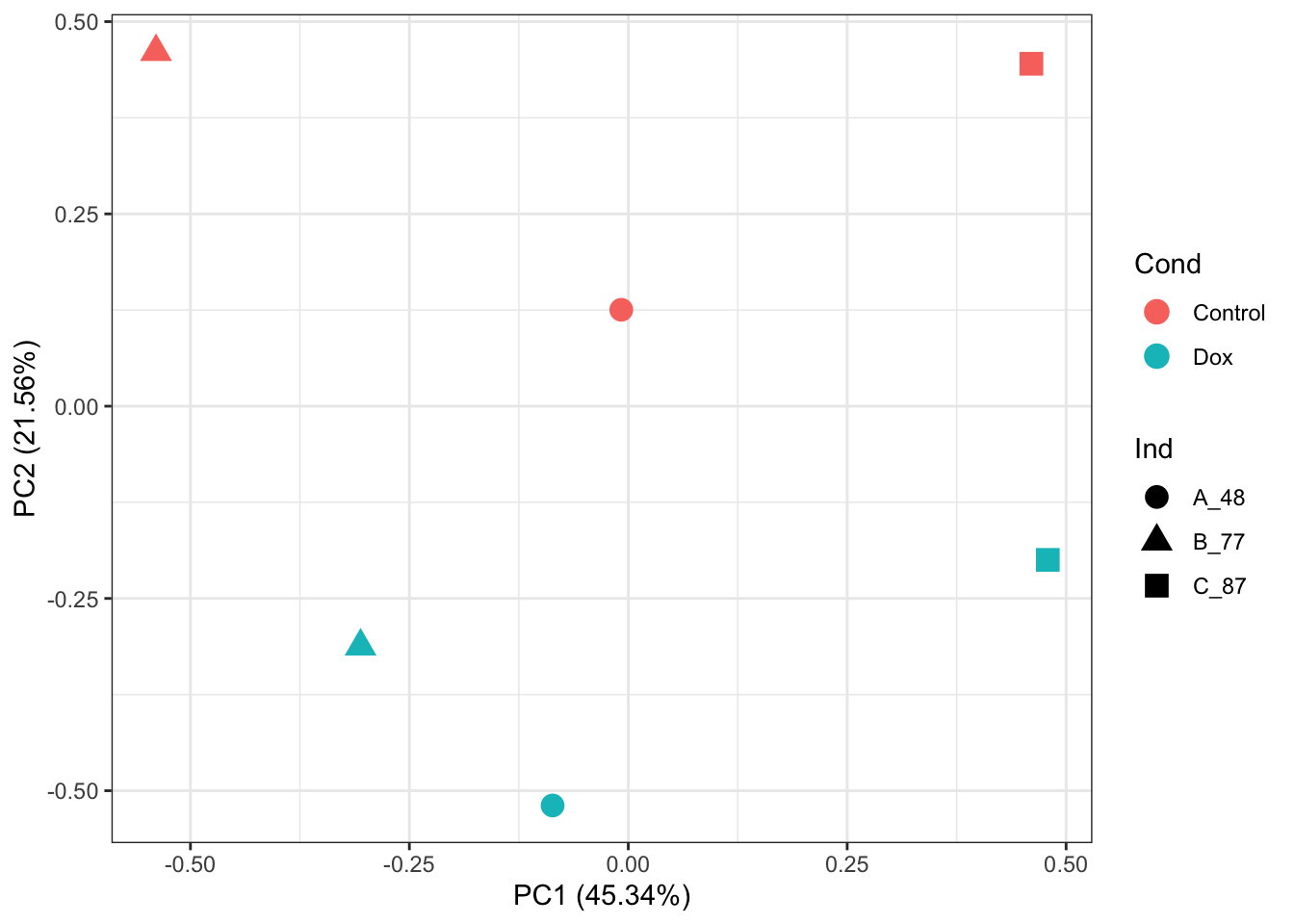
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Volcano plots
# 1. Create a column to threshold P-values
toptable_summary <- toptable_summary %>% mutate(threshold_P = P.Value < 0.05)
# 2. Plot
ggplot(toptable_summary)+
geom_point(mapping = aes(x = logFC, y = -log10(P.Value), color = threshold_P))+
xlab("log2FC")+
ylab("-log10 nominal p-value")+
ylim(0, 10)+
xlim(-7, 7)+
theme(legend.position = "none",
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = rel(1.25)))+
theme_bw()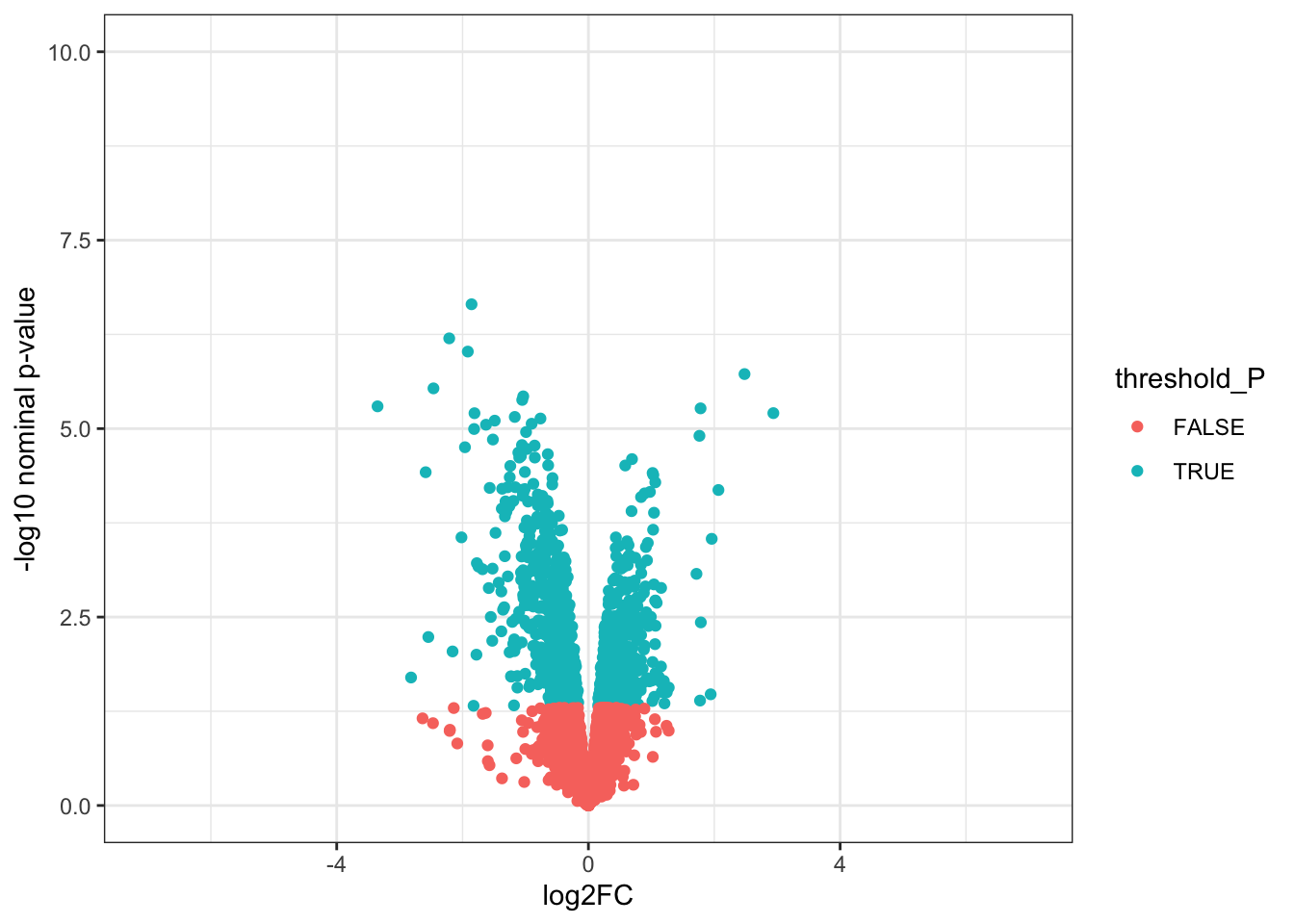
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Make top_proteins your toptable summary
top_proteins <- toptable_summary[1:5, ] %>% rownames()
# Loop through each of the top proteins within the Model_counts
for (i in top_proteins) {
# Create a boxplot for each protein
boxplot(counts[i, ] ~ phenoData_sub$Cond,
main = i,
xlab = "Condition",
ylab = "log2-RUVg-Quantnorm Abundance",
col = c("lightblue", "red2"),
las = 2) # las = 2 makes the axis labels perpendicular to the axis
}
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Linear model + RUVg + Imputation
# Convert to DGEList object
dge <- DGEList(counts = as.matrix(Protein_DF_Abundance_imp_2))
# Calculate variability
cv <- rowSds(dge$counts, useNames = TRUE)/rowMeans(dge$counts)
# Identify the 10% least variable proteins
num_controls <- ceiling(0.1 * nrow(dge$counts))
control_proteins <- names(sort(cv, decreasing = FALSE))[1:num_controls]
control_proteins %>% length()[1] 419# Round abundance values (applies more specifically to the imputed values) so that the abundance matrix is valid input into newSeqExpressionSet
dge$counts <- round(dge$counts)
dge$counts %>% dim()[1] 4182 10# Generate newSeqExpressionSet object
dds<- newSeqExpressionSet(counts = dge$counts, phenoData = Meta )
# Apply RUVg normalization
set_ruv <- RUVg(dds, control_proteins, k = 1)
# View the normalized counts from RUVg
normCounts(set_ruv) %>% head() S1 S3 S5 S7 S9 S2 S4
A0A0B4J2A2 1170994 1177013 1594300 1671732 1807304 1162508 1412564
A0A0B4J2D5 15635010 17496725 15620877 14271514 15377563 14885677 14140379
A0A494C071 418258 382940 300651 427346 392772 483809 448568
A0AVT1 79578 100870 178768 134888 134312 65444 90070
A0FGR8 1243016 1116610 970294 1040982 1020852 1123241 1114550
A0JLT2 214334 185260 181142 144459 207455 220417 209395
S6 S8 S10
A0A0B4J2A2 1480424 1388010 1555148
A0A0B4J2D5 13575542 13202450 12666875
A0A494C071 379025 460238 348243
A0AVT1 129863 84523 96901
A0FGR8 1080205 1119978 1059410
A0JLT2 159940 126144 210538# PCA of RUVg normalized abundance values:
prcomp_res <- prcomp(t(normCounts(set_ruv) %>% as.matrix()), center = TRUE, scale. = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(Meta), colour = "Cond", shape = "Ind", size =4)+
theme_bw()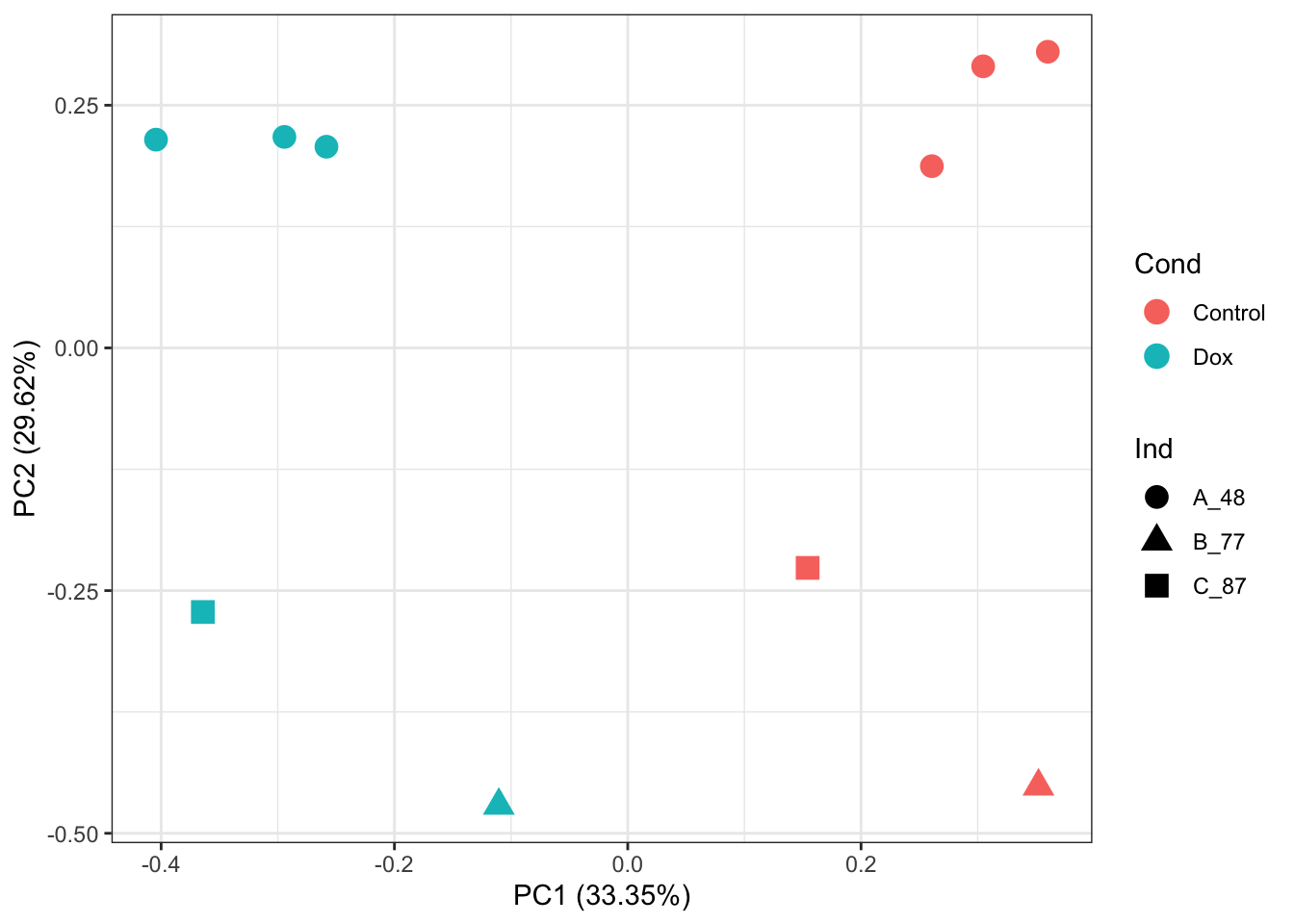
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2 <- normCounts(set_ruv) %>% log2()
counts_RUVg_log2 %>% head() S1 S3 S5 S7 S9 S2 S4
A0A0B4J2A2 20.15930 20.16670 20.60449 20.67291 20.78541 20.14881 20.42988
A0A0B4J2D5 23.89828 24.06058 23.89697 23.76664 23.87432 23.82742 23.75332
A0A494C071 18.67403 18.54676 18.19773 18.70505 18.58333 18.88408 18.77497
A0AVT1 16.28008 16.62214 17.44773 17.04140 17.03523 15.99797 16.45876
A0FGR8 20.24541 20.09069 19.88806 19.98951 19.96134 20.09924 20.08803
A0JLT2 17.70950 17.49919 17.46676 17.14030 17.66244 17.74988 17.67587
S6 S8 S10
A0A0B4J2A2 20.49758 20.40459 20.56862
A0A0B4J2D5 23.69451 23.65430 23.59456
A0A494C071 18.53193 18.81202 18.40973
A0AVT1 16.98663 16.36706 16.56422
A0FGR8 20.04287 20.09504 20.01483
A0JLT2 17.28717 16.94471 17.68372counts_RUVg_log2 %>% dim()[1] 4182 10prcomp_res <- prcomp(t(counts_RUVg_log2 %>% as.matrix()), center = TRUE, scale. = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(Meta), colour = "Cond", shape = "Ind", size =4)+
theme_bw()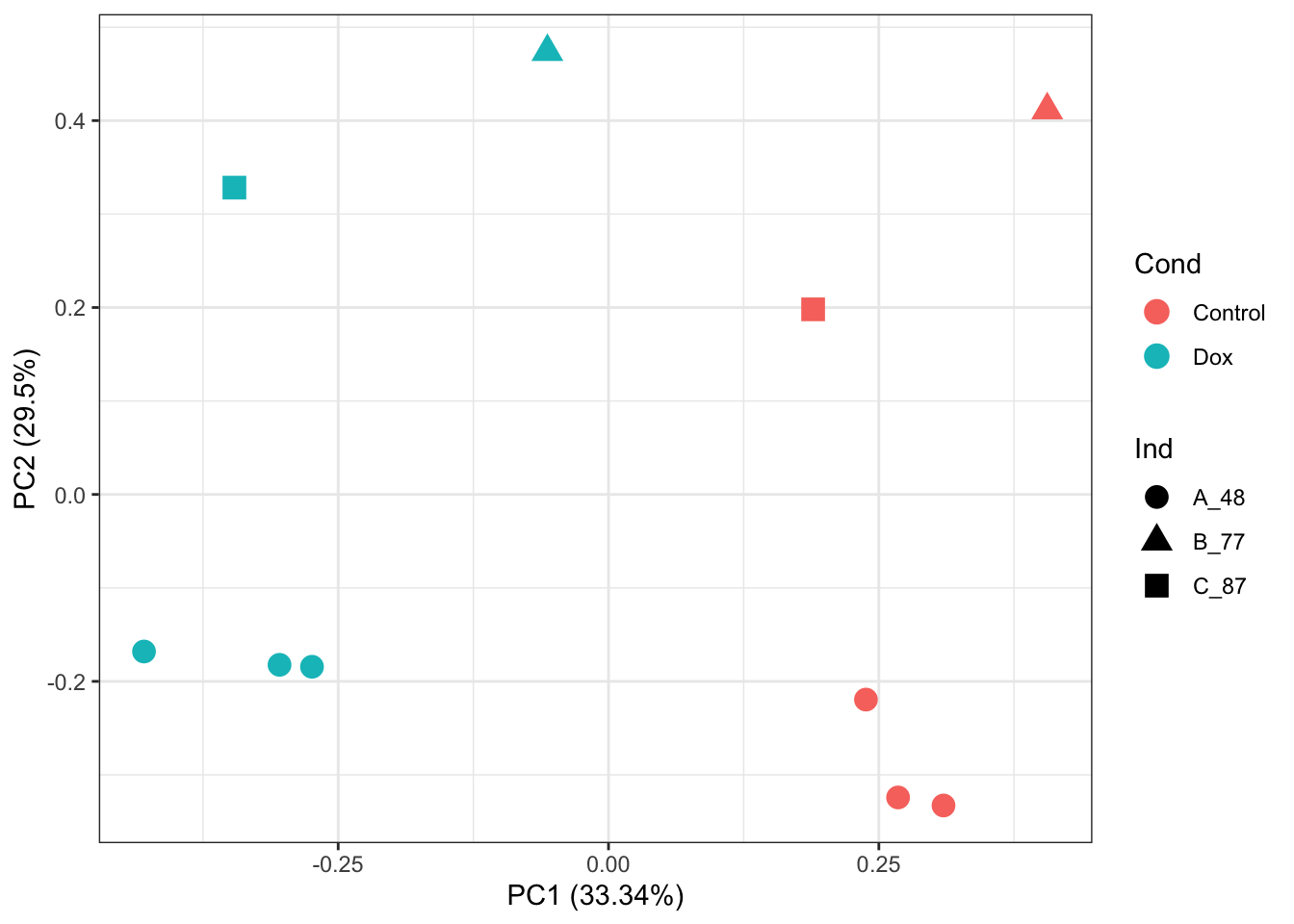
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2[,1] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2[,3] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2[,4] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2[,5] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2[,6] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2[,7] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2[,8] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2[,9] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2[,10] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Meta Samples Ind Rep Cond Cond_Ind
S1 S1 B_77 Bio Dox B_77_Dox
S3 S3 C_87 Bio Dox C_87_Dox
S5 S5 A_48 Tech Dox A_48_Dox
S7 S7 A_48 Tech Dox A_48_Dox
S9 S9 A_48 Tech Dox A_48_Dox
S2 S2 B_77 Bio Control B_77_Control
S4 S4 C_87 Bio Control C_87_Control
S6 S6 A_48 Tech Control A_48_Control
S8 S8 A_48 Tech Control A_48_Control
S10 S10 A_48 Tech Control A_48_ControlphenoData_sub <- Meta[c(1,2,4,6,7,9),]
counts_RUVg_log2_sub <- counts_RUVg_log2[,c(1,2,4,6,7,9)]
counts_RUVg_log2_sub %>% head() S1 S3 S7 S2 S4 S8
A0A0B4J2A2 20.15930 20.16670 20.67291 20.14881 20.42988 20.40459
A0A0B4J2D5 23.89828 24.06058 23.76664 23.82742 23.75332 23.65430
A0A494C071 18.67403 18.54676 18.70505 18.88408 18.77497 18.81202
A0AVT1 16.28008 16.62214 17.04140 15.99797 16.45876 16.36706
A0FGR8 20.24541 20.09069 19.98951 20.09924 20.08803 20.09504
A0JLT2 17.70950 17.49919 17.14030 17.74988 17.67587 16.94471counts_RUVg_log2_sub %>% dim()[1] 4182 6prcomp_res <- prcomp(t(counts_RUVg_log2_sub %>% as.matrix()), center = TRUE, scale. = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(phenoData_sub), colour = "Cond", shape = "Ind", size =4)+
theme_bw()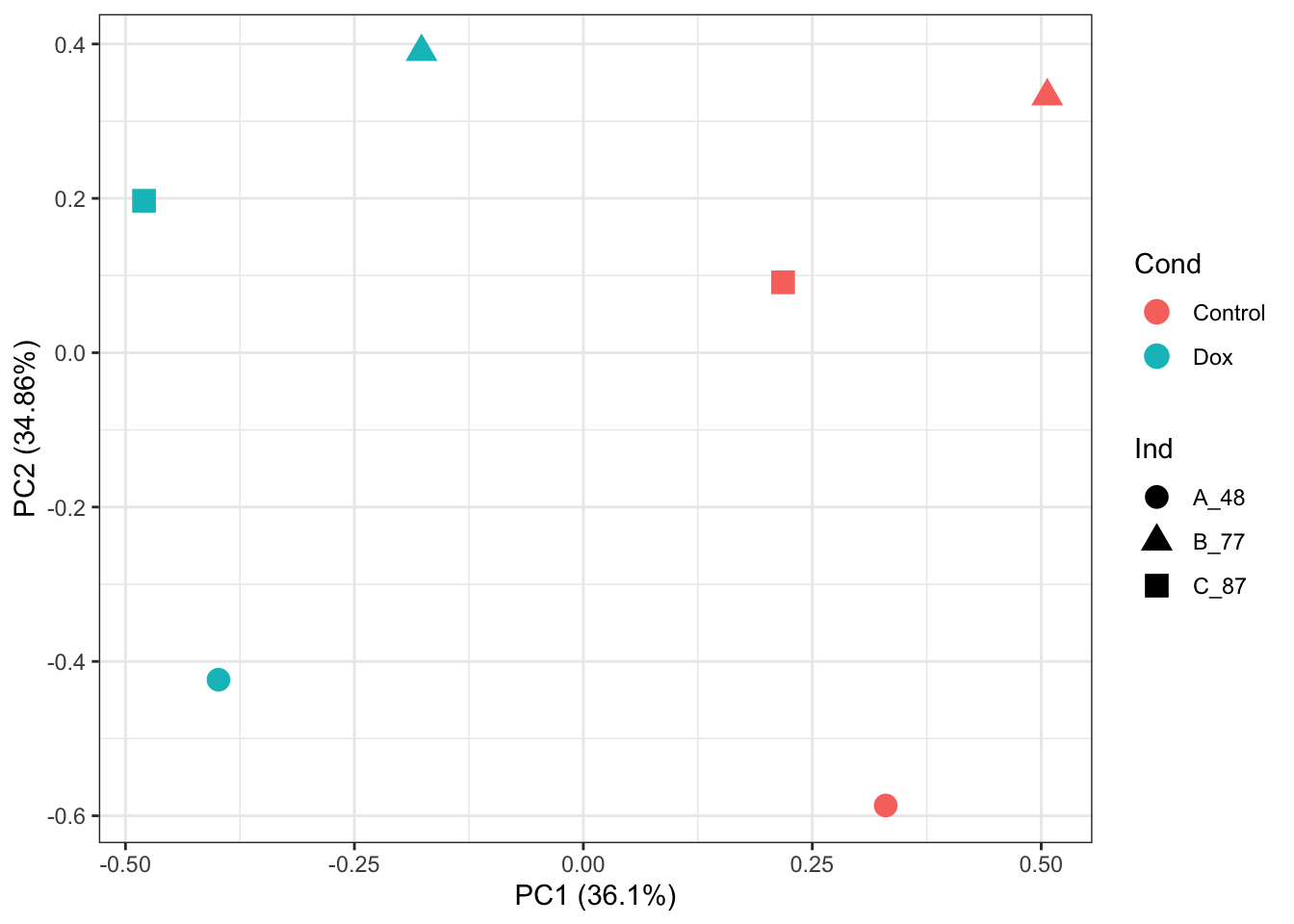
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2_sub[,1] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2_sub[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2_sub[,3] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2_sub[,4] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2_sub[,5] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2_sub[,6] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Quantile normalization after RUVg normalized abundances have been
# subset to 6 samples and log2 transformed
counts_RUVg_log2_sub_quantnorm <- limma::normalizeBetweenArrays(counts_RUVg_log2_sub, method = "quantile")
counts_RUVg_log2_sub_quantnorm %>% head() S1 S3 S7 S2 S4 S8
A0A0B4J2A2 20.13888 20.23864 20.67329 20.16485 20.42308 20.31020
A0A0B4J2D5 23.90579 24.01114 23.76820 23.82365 23.81224 23.51349
A0A494C071 18.65403 18.67200 18.76198 18.83975 18.72029 18.74987
A0AVT1 16.36156 16.80301 17.05071 15.91893 16.26320 16.31186
A0FGR8 20.22776 20.16953 20.01610 20.10131 20.08928 20.00796
A0JLT2 17.70234 17.63727 17.19631 17.68290 17.57631 16.89455counts_RUVg_log2_sub_quantnorm %>% dim()[1] 4182 6# PCA of log2-quantile normalized-RUVs values:
prcomp_res <- prcomp(t(counts_RUVg_log2_sub_quantnorm %>% as.matrix()), center = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(phenoData_sub), colour = "Cond", shape = "Ind", size =4)+
theme_bw()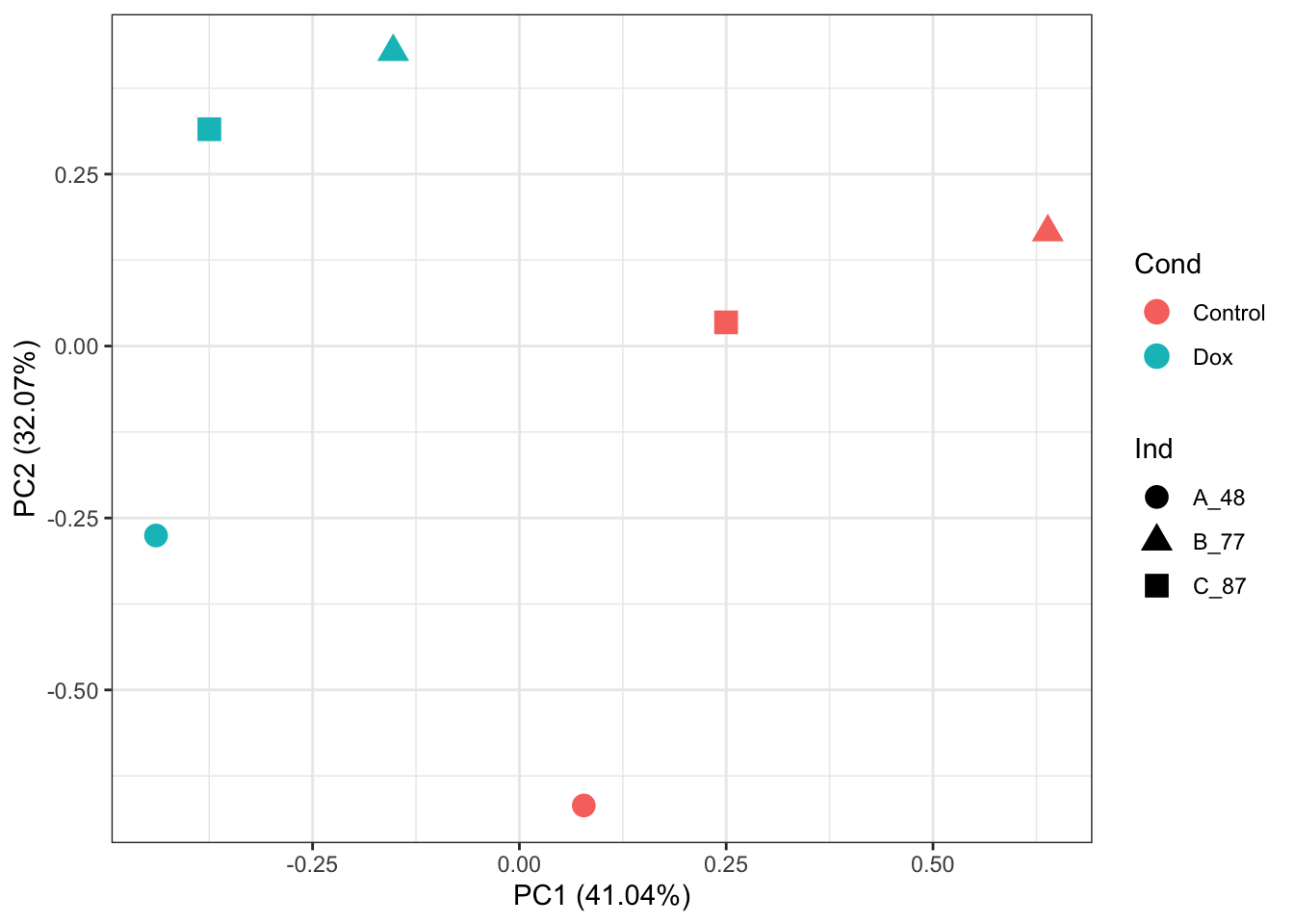
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2_sub_quantnorm[,1] %>% hist(breaks = 100)
counts_RUVg_log2_sub_quantnorm[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUVg_log2_sub_quantnorm[,3] %>% hist(breaks = 100)
counts_RUVg_log2_sub_quantnorm[,4] %>% hist(breaks = 100)
counts_RUVg_log2_sub_quantnorm[,5] %>% hist(breaks = 100)
counts_RUVg_log2_sub_quantnorm[,6] %>% hist(breaks = 100)
# Create a DataFrame for the phenoData
phenoData_RUVg <- DataFrame(Meta)
phenoData_RUVg <- Meta[c(1,2,4,6,7,9), ]
set_ruv$W_1 [1] -0.341427911 0.426122666 -0.168696007 -0.132609568 -0.117337568
[6] -0.406192248 0.685705127 -0.026632222 0.002746413 0.078321318RUV_1 <- set_ruv$W_1
phenoData_RUVg$RUV_1 <- RUV_1[c(1,2,4,6,7,9)]
# View changes
phenoData_RUVg %>% head() Samples Ind Rep Cond Cond_Ind RUV_1
S1 S1 B_77 Bio Dox B_77_Dox -0.341427911
S3 S3 C_87 Bio Dox C_87_Dox 0.426122666
S7 S7 A_48 Tech Dox A_48_Dox -0.132609568
S2 S2 B_77 Bio Control B_77_Control -0.406192248
S4 S4 C_87 Bio Control C_87_Control 0.685705127
S8 S8 A_48 Tech Control A_48_Control 0.002746413phenoData_RUVg Samples Ind Rep Cond Cond_Ind RUV_1
S1 S1 B_77 Bio Dox B_77_Dox -0.341427911
S3 S3 C_87 Bio Dox C_87_Dox 0.426122666
S7 S7 A_48 Tech Dox A_48_Dox -0.132609568
S2 S2 B_77 Bio Control B_77_Control -0.406192248
S4 S4 C_87 Bio Control C_87_Control 0.685705127
S8 S8 A_48 Tech Control A_48_Control 0.002746413# Create Design Matrix
# phenoData$Cond <- factor(phenoData$Cond , levels = c("Control", "Dox"))
design <- model.matrix(~ 0 + Cond + RUV_1, data = phenoData_RUVg)
design CondControl CondDox RUV_1
S1 0 1 -0.341427911
S3 0 1 0.426122666
S7 0 1 -0.132609568
S2 1 0 -0.406192248
S4 1 0 0.685705127
S8 1 0 0.002746413
attr(,"assign")
[1] 1 1 2
attr(,"contrasts")
attr(,"contrasts")$Cond
[1] "contr.treatment"# rename columns
colnames(design) <- c('Control', "Dox", "RUV_1")
# Get model counts
Model_counts <- counts_RUVg_log2_sub_quantnorm
# Fit model
dupcor <- duplicateCorrelation(Model_counts, design = design, block = phenoData_RUVg$Ind)
fit <- lmFit(object = as.matrix(Model_counts), block = phenoData_RUVg$Ind, design = design, correlation = dupcor$consensus.correlation)
fit2 <- eBayes(fit)
# Make contrasts
cm <- makeContrasts(
DoxvNorm = Dox - Control,
RUV1vNorm = RUV_1 - Control,
levels = design)
# Model with contrasts
fit2 <- contrasts.fit(fit, cm)
fit2 <- eBayes(fit2, robust = TRUE, trend = TRUE)Warning: 4 very small variances detected, have been offset away from zero# Summarize
results_summary <- decideTests(fit2, adjust.method = "none", p.value = 0.05)
summary(results_summary) DoxvNorm RUV1vNorm
Down 536 4182
NotSig 3019 0
Up 627 0# Toptable summary organized
toptable_summary_DoxvNorm <- topTable(fit2, coef = "DoxvNorm",number = (nrow(Model_counts)), p.value = 1, adjust.method = "none")
toptable_summary_DoxvNorm$Protein <- rownames(toptable_summary_DoxvNorm)
toptable_summary_DoxvNorm$P.Value %>% hist(breaks = 100)
abline(v = c(0.01,0.025, 0.05, 0.1), col = "red" )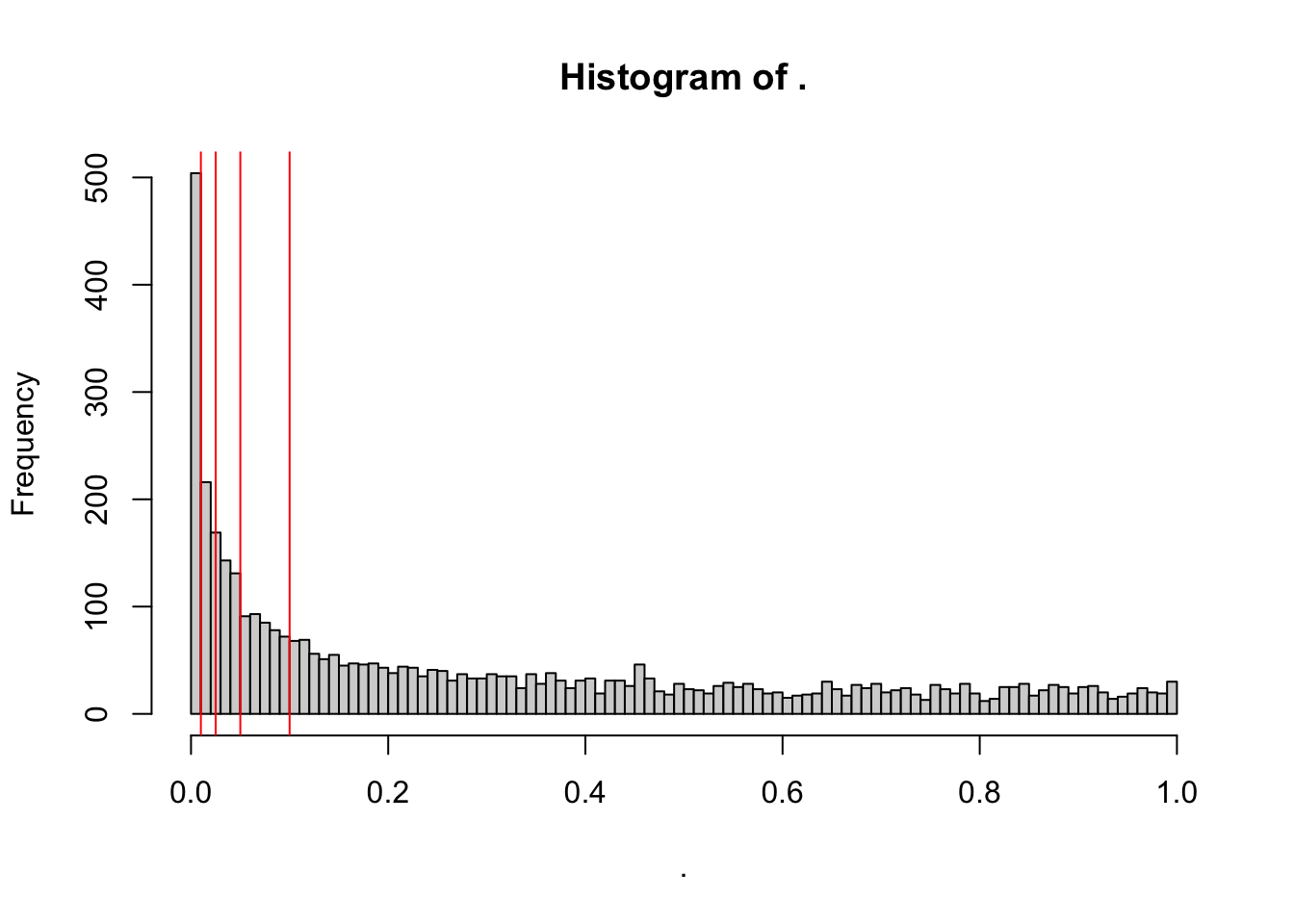
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Volcano plots
# 1. Create a column to threshold P-values
toptable_summary_DoxvNorm <- toptable_summary_DoxvNorm %>% mutate(threshold_P = P.Value < 0.05)
# 2. Plot
ggplot(toptable_summary_DoxvNorm)+
geom_point(mapping = aes(x = logFC, y = -log10(P.Value), color = threshold_P))+
xlab("log2FC")+
ylab("-log10 nominal p-value")+
ylim(0, 6)+
xlim(-6, 6)+
theme(legend.position = "none",
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = rel(1.25)))+
theme_bw()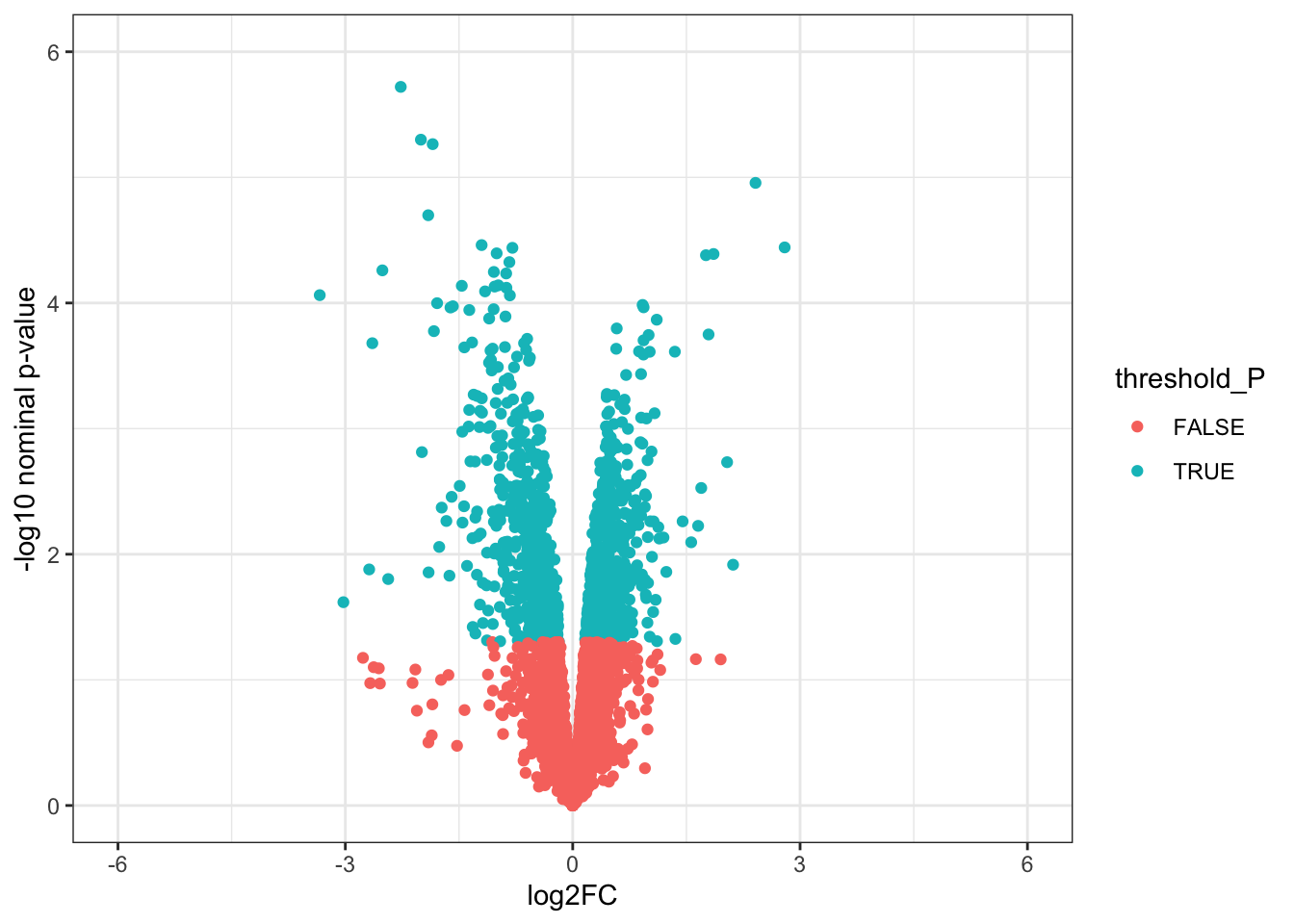
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Make top_proteins your toptable summary
top_proteins <- toptable_summary_DoxvNorm[1:5, ] %>% rownames()
# Loop through each of the top proteins within the Model_counts
for (i in top_proteins) {
# Create a boxplot for each protein
boxplot(Model_counts[i, ] ~ phenoData_RUVg$Cond,
main = i,
xlab = "Condition",
ylab = "log2-RUVg-Quantnorm Abundance",
col = c("lightblue", "red2"),
las = 2) # las = 2 makes the axis labels perpendicular to the axis
}
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
FigS2 C&D RUV III
# Convert to DGEList object
dge <- DGEList(counts = as.matrix(Protein_DF_Abundance_imp_2))
# Calculate variability
cv <- rowSds(dge$counts, useNames = TRUE)/rowMeans(dge$counts)
# Identify the 10% least variable proteins
num_controls <- ceiling(0.05 * nrow(dge$counts))
control_proteins <- names(sort(cv, decreasing = FALSE))[1:num_controls]
control_proteins %>% length()[1] 210# Create an index vector
control_proteins_index <- match(control_proteins, rownames(Protein_DF_Abundance_imp_2))
control_proteins_index %>% length()[1] 210# Design matrix from metadata
Meta Samples Ind Rep Cond Cond_Ind
S1 S1 B_77 Bio Dox B_77_Dox
S3 S3 C_87 Bio Dox C_87_Dox
S5 S5 A_48 Tech Dox A_48_Dox
S7 S7 A_48 Tech Dox A_48_Dox
S9 S9 A_48 Tech Dox A_48_Dox
S2 S2 B_77 Bio Control B_77_Control
S4 S4 C_87 Bio Control C_87_Control
S6 S6 A_48 Tech Control A_48_Control
S8 S8 A_48 Tech Control A_48_Control
S10 S10 A_48 Tech Control A_48_ControlModel_mat <- replicate.matrix(a = Meta[,c(4)])
Model_mat Control Dox
1 0 1
2 0 1
3 0 1
4 0 1
5 0 1
6 1 0
7 1 0
8 1 0
9 1 0
10 1 0
attr(,"assign")
[1] 1 1
attr(,"contrasts")
attr(,"contrasts")$a
[1] "contr.treatment"Test <- RUVIII(Y = t(Protein_DF_Abundance_imp_2 %>% log2()), M = Model_mat, ctl =control_proteins_index , k = 1, return.info = TRUE)
New_Y <- Test$newY %>% t()
New_Y[(New_Y %>% rowMeans()) < 0, ] S1 S3 S5 S7 S9 S2 S4 S6 S8 S10# PCA of Pre-RUVIII normalized abundance values:
prcomp_res <- prcomp(t(Protein_DF_Abundance_imp_2 %>% as.matrix()), center = TRUE, scale. = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(Meta), colour = "Cond", shape = "Ind", size =4)+
theme_bw()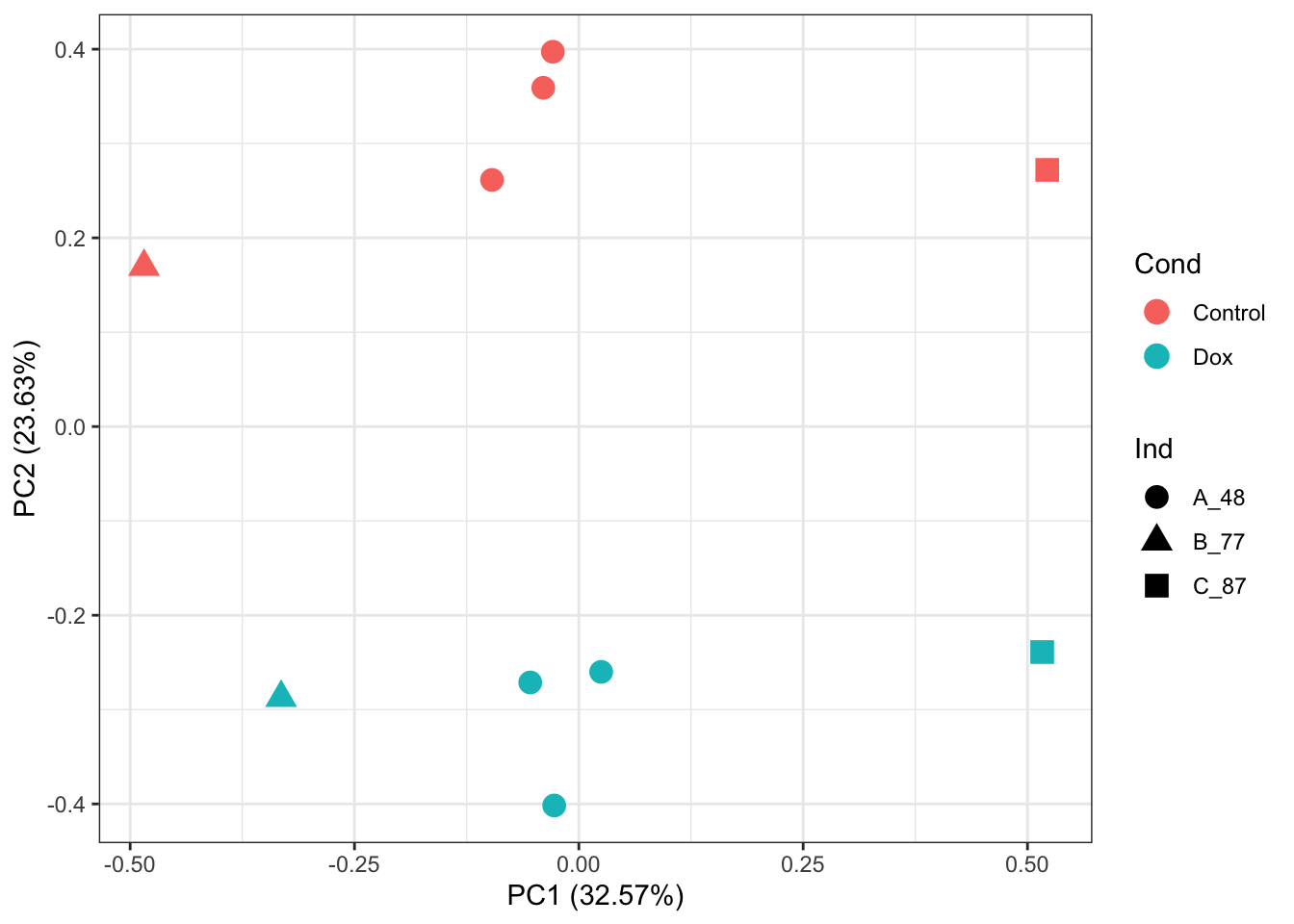
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# PCA of Post-RUVIII normalized abundance values:
prcomp_res <- prcomp(t(New_Y %>% as.matrix()), center = TRUE, scale. = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(Meta), colour = "Cond", shape = "Ind", size =4)+
theme_bw()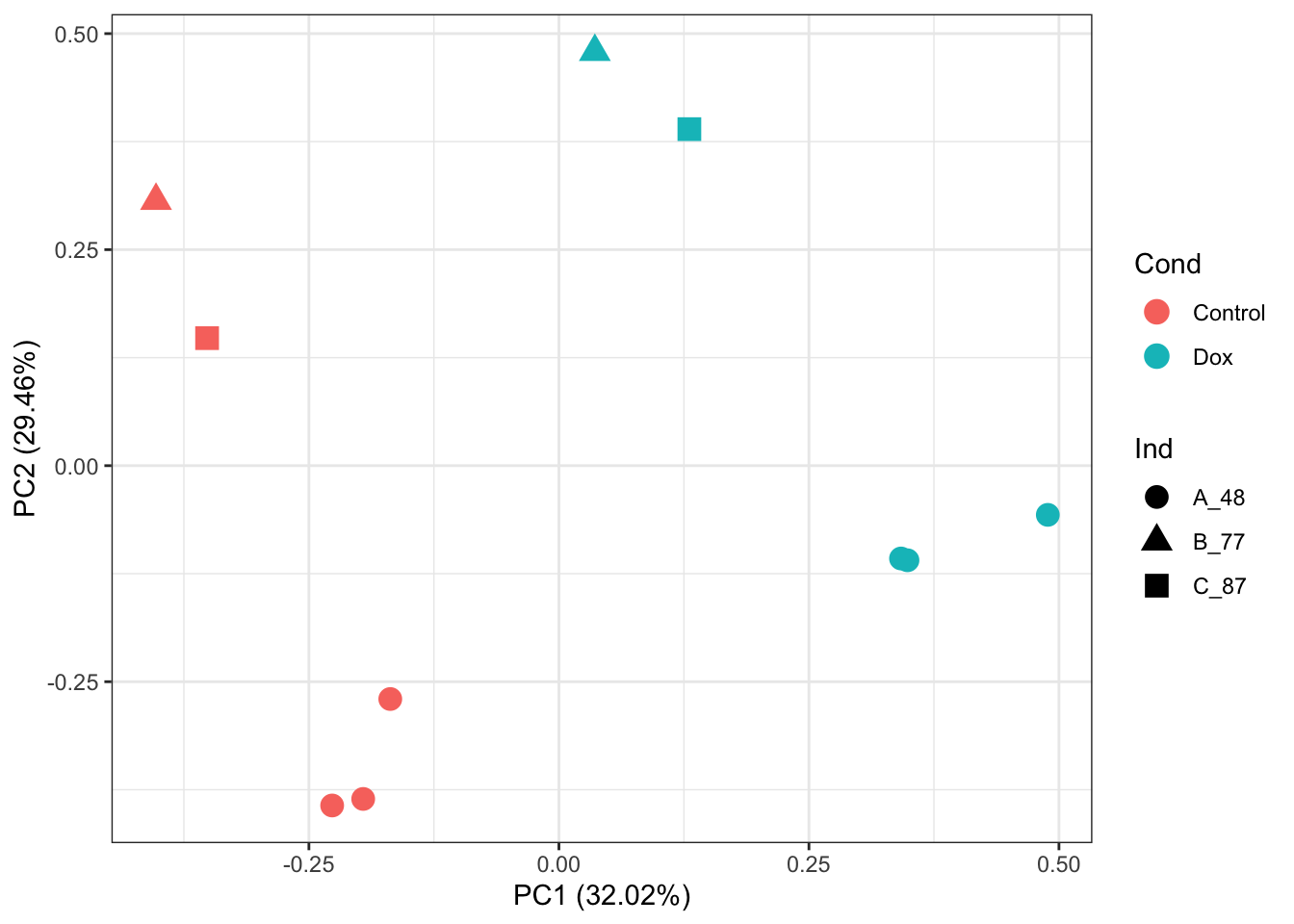
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y %>% head() S1 S3 S5 S7 S9 S2 S4
A0A0B4J2A2 18.83632 18.79443 19.28942 19.40656 19.49348 18.97978 19.24259
A0A0B4J2D5 23.96540 24.07276 23.95113 23.81659 23.92396 23.89443 23.74051
A0A494C071 18.14255 18.08070 17.68807 18.21752 18.08793 18.40303 18.40635
A0AVT1 14.29941 14.44480 15.45203 15.11493 15.06703 14.26425 14.52497
A0FGR8 20.26983 20.13941 19.91745 20.01871 19.99166 20.11768 20.13968
A0JLT2 17.90073 17.73392 17.66481 17.33240 17.85925 17.91398 17.89374
S6 S8 S10
A0A0B4J2A2 19.25256 19.16810 19.31979
A0A0B4J2D5 23.73598 23.69334 23.62843
A0A494C071 18.06396 18.35044 17.95179
A0AVT1 15.07628 14.46519 14.63148
A0FGR8 20.07468 20.12751 20.04989
A0JLT2 17.48087 17.13845 17.88291New_Y %>% rowMeans() %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y %>% na.omit() %>% dim()[1] 4182 10New_Y[,1] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y[,3] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y[,4] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y[,5] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y[,6] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y[,7] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y[,8] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y[,9] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
New_Y[,10] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
Meta_sub<-Meta[c(1,2,4,6,7,9),]
counts_RUV_log2_sub <- New_Y[,c(1,2,4,6,7,9)]
counts_RUV_log2_sub %>% head() S1 S3 S7 S2 S4 S8
A0A0B4J2A2 18.83632 18.79443 19.40656 18.97978 19.24259 19.16810
A0A0B4J2D5 23.96540 24.07276 23.81659 23.89443 23.74051 23.69334
A0A494C071 18.14255 18.08070 18.21752 18.40303 18.40635 18.35044
A0AVT1 14.29941 14.44480 15.11493 14.26425 14.52497 14.46519
A0FGR8 20.26983 20.13941 20.01871 20.11768 20.13968 20.12751
A0JLT2 17.90073 17.73392 17.33240 17.91398 17.89374 17.13845counts_RUV_log2_sub %>% dim()[1] 4182 6counts_RUV_log2_sub[,1] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUV_log2_sub[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUV_log2_sub[,3] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUV_log2_sub[,4] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUV_log2_sub[,5] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUV_log2_sub[,6] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# PCA of RUVIII normalized abundance values:
prcomp_res <- prcomp(t(counts_RUV_log2_sub %>% as.matrix()), center = TRUE, scale. = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(Meta_sub), colour = "Cond", shape = "Ind", size =4)+
theme_bw()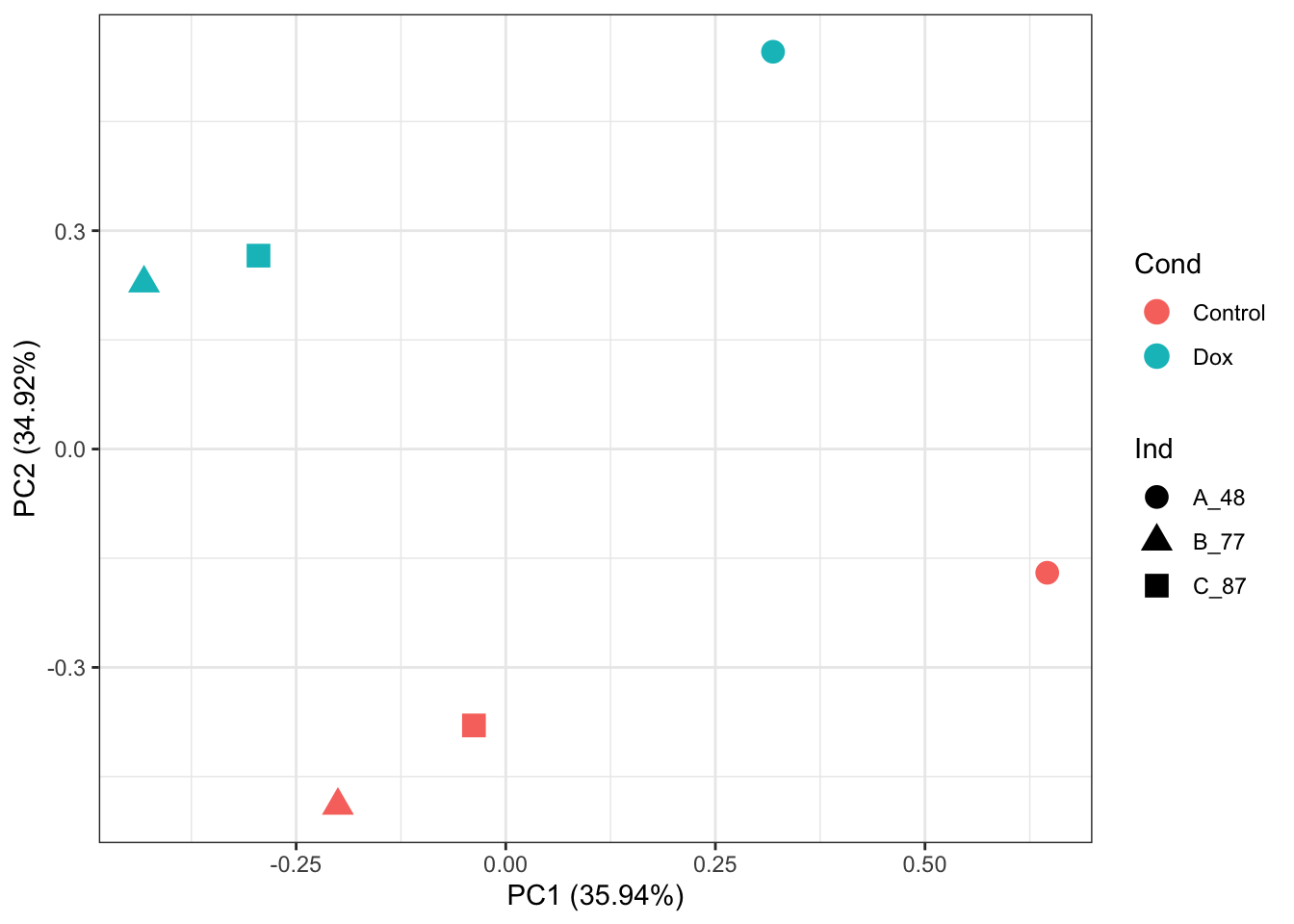
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUV_log2_sub_quantnorm <- limma::normalizeBetweenArrays(counts_RUV_log2_sub, method = "quantile")
counts_RUV_log2_sub_quantnorm %>% head() S1 S3 S7 S2 S4 S8
A0A0B4J2A2 18.86297 18.90679 19.42470 18.98217 19.19023 19.07789
A0A0B4J2D5 23.92284 24.05312 23.80434 23.96008 23.78608 23.61631
A0A494C071 18.18276 18.18276 18.23292 18.39560 18.33624 18.28836
A0AVT1 14.39705 14.50740 15.22635 14.27202 14.39705 14.42666
A0FGR8 20.25839 20.18068 20.05801 20.12714 20.10611 20.06667
A0JLT2 17.92053 17.84082 17.39529 17.88697 17.81814 17.05696counts_RUV_log2_sub_quantnorm %>% dim()[1] 4182 6counts_RUV_log2_sub_quantnorm[,1] %>% hist(breaks = 100)
counts_RUV_log2_sub_quantnorm[,2] %>% hist(breaks = 100)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
counts_RUV_log2_sub_quantnorm[,3] %>% hist(breaks = 100)
counts_RUV_log2_sub_quantnorm[,4] %>% hist(breaks = 100)
counts_RUV_log2_sub_quantnorm[,5] %>% hist(breaks = 100)
counts_RUV_log2_sub_quantnorm[,6] %>% hist(breaks = 100)
# PCA of RUVIII normalized abundance values:
prcomp_res <- prcomp(t(counts_RUV_log2_sub_quantnorm %>% as.matrix()), center = TRUE, scale. = TRUE)
ggplot2::autoplot(prcomp_res, data = as.data.frame(Meta_sub), colour = "Cond", shape = "Ind", size =4)+
theme_bw()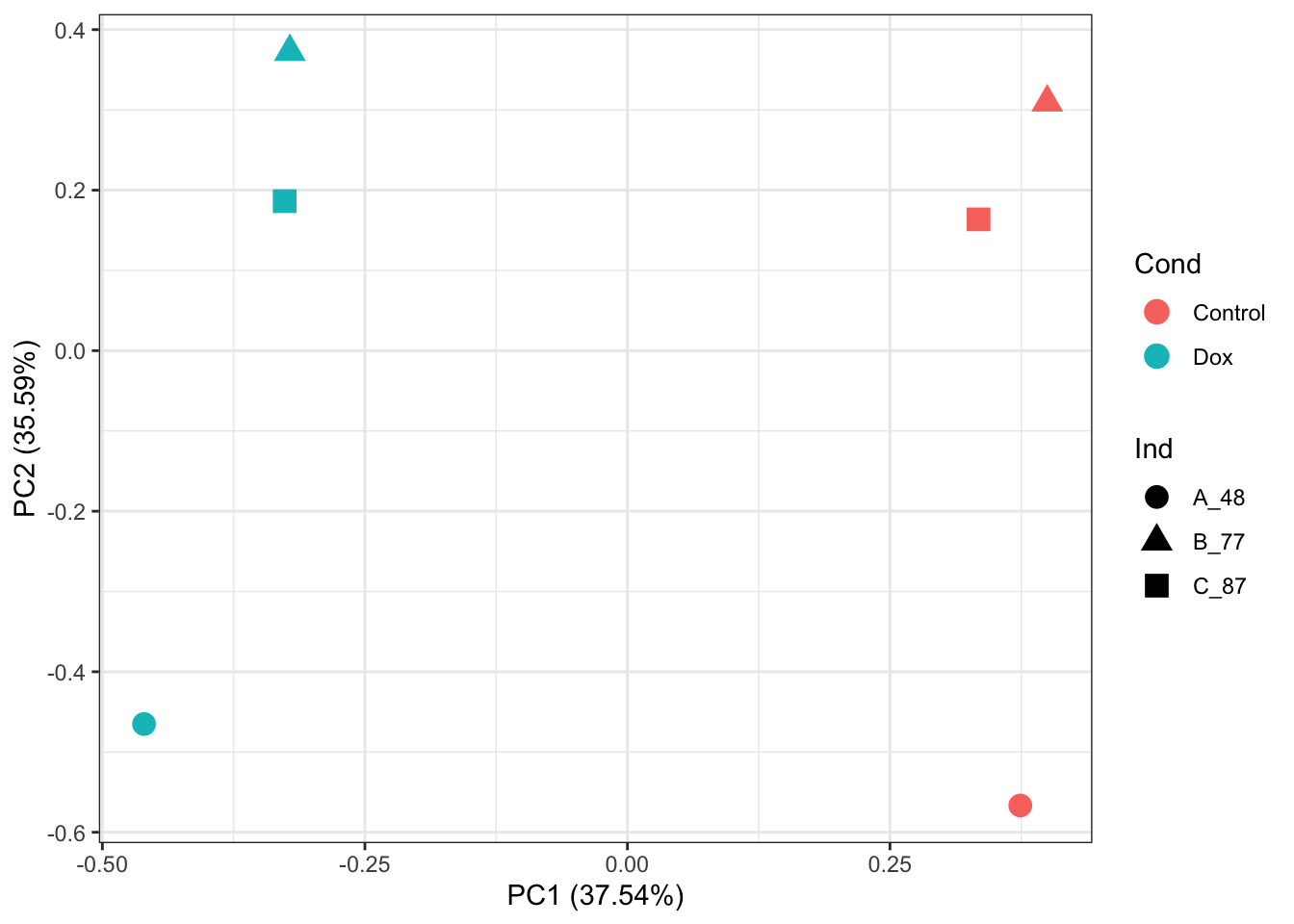
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Create a DataFrame for the phenoData
phenoData_RUVIII <- DataFrame(Meta)
phenoData_RUVIII <- Meta[c(1,2,4,6,7,9), ]
# View changes
phenoData_RUVIII Samples Ind Rep Cond Cond_Ind
S1 S1 B_77 Bio Dox B_77_Dox
S3 S3 C_87 Bio Dox C_87_Dox
S7 S7 A_48 Tech Dox A_48_Dox
S2 S2 B_77 Bio Control B_77_Control
S4 S4 C_87 Bio Control C_87_Control
S8 S8 A_48 Tech Control A_48_Control# Create Design Matrix
# phenoData$Cond <- factor(phenoData$Cond , levels = c("Control", "Dox"))
design <- model.matrix(~ 0 + Cond, data = phenoData_RUVIII)
design CondControl CondDox
S1 0 1
S3 0 1
S7 0 1
S2 1 0
S4 1 0
S8 1 0
attr(,"assign")
[1] 1 1
attr(,"contrasts")
attr(,"contrasts")$Cond
[1] "contr.treatment"# rename columns
colnames(design) <- c('Control', "Dox")
# Get model counts
Model_counts <- counts_RUV_log2_sub_quantnorm
# Fit model
dupcor <- duplicateCorrelation(Model_counts, design = design, block = phenoData_RUVIII$Ind)
fit <- lmFit(object = as.matrix(Model_counts), block = phenoData_RUVIII$Ind, design = design, correlation = dupcor$consensus.correlation)
fit2 <- eBayes(fit)
# Make contrasts
cm <- makeContrasts(
DoxvNorm = Dox - Control,
levels = design)
# Model with contrasts
fit2 <- contrasts.fit(fit, cm)
fit2 <- eBayes(fit2, robust = TRUE, trend = TRUE)
# Summarize
results_summary <- decideTests(fit2, adjust.method = "none", p.value = 0.05)
summary(results_summary) DoxvNorm
Down 546
NotSig 3027
Up 609# Toptable summary organized
toptable_summary_DoxvNorm <- topTable(fit2, coef = "DoxvNorm",number = (nrow(Model_counts)), p.value = 1, adjust.method = "none")
toptable_summary_DoxvNorm$Protein <- rownames(toptable_summary_DoxvNorm)
toptable_summary_DoxvNorm$P.Value %>% hist(breaks = 100)
abline(v = c(0.01,0.025, 0.05, 0.1), col = "red" )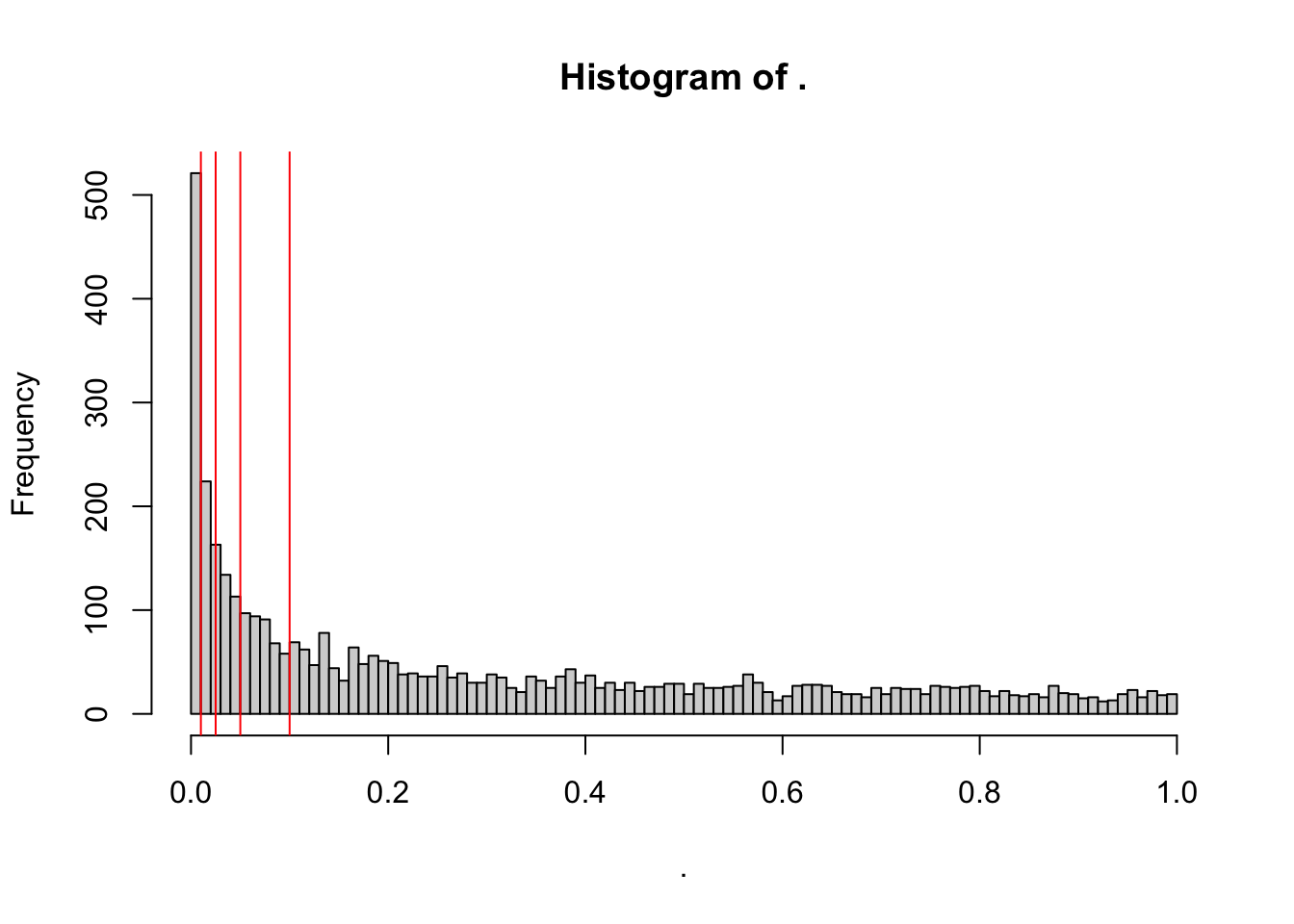
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
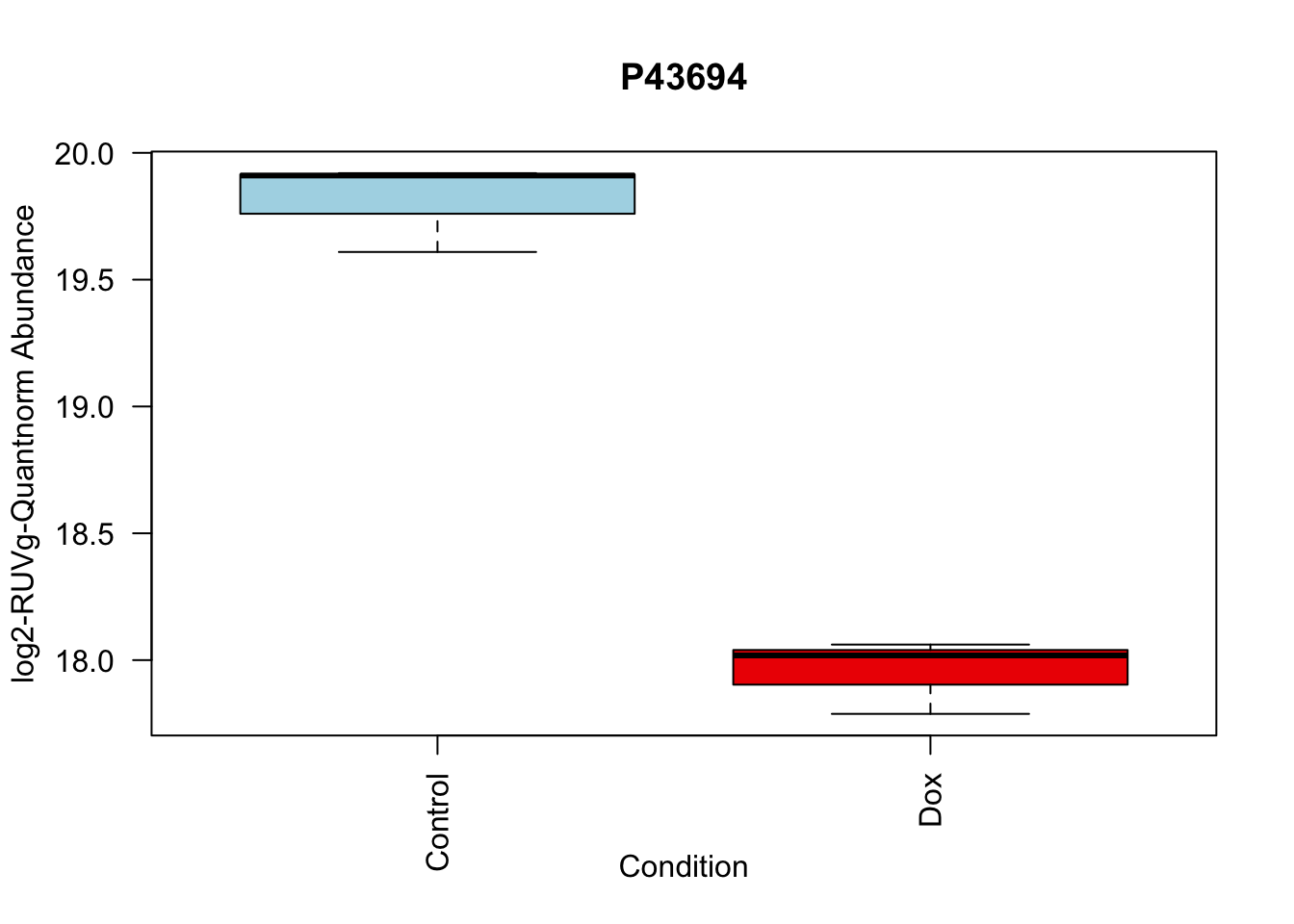
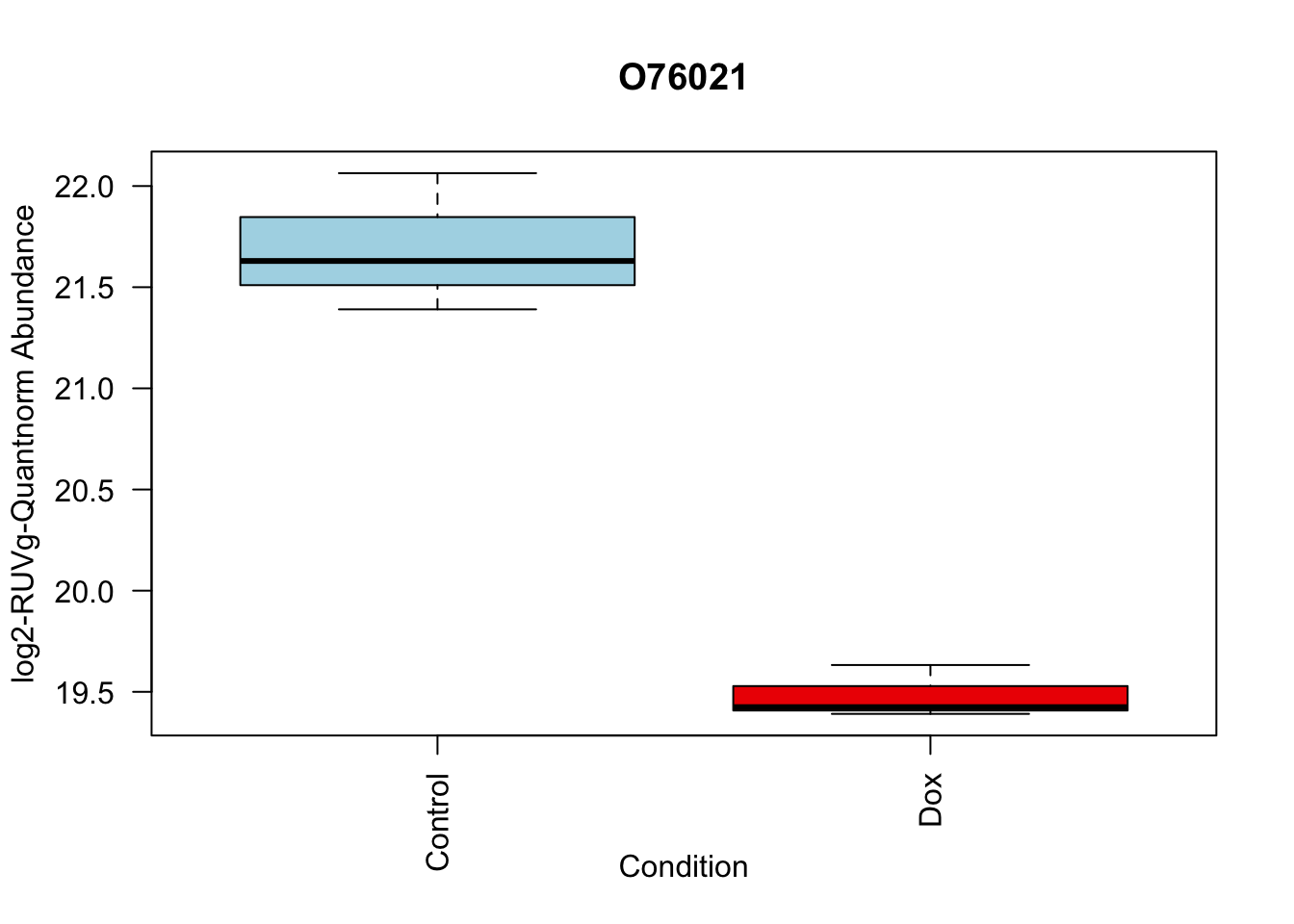
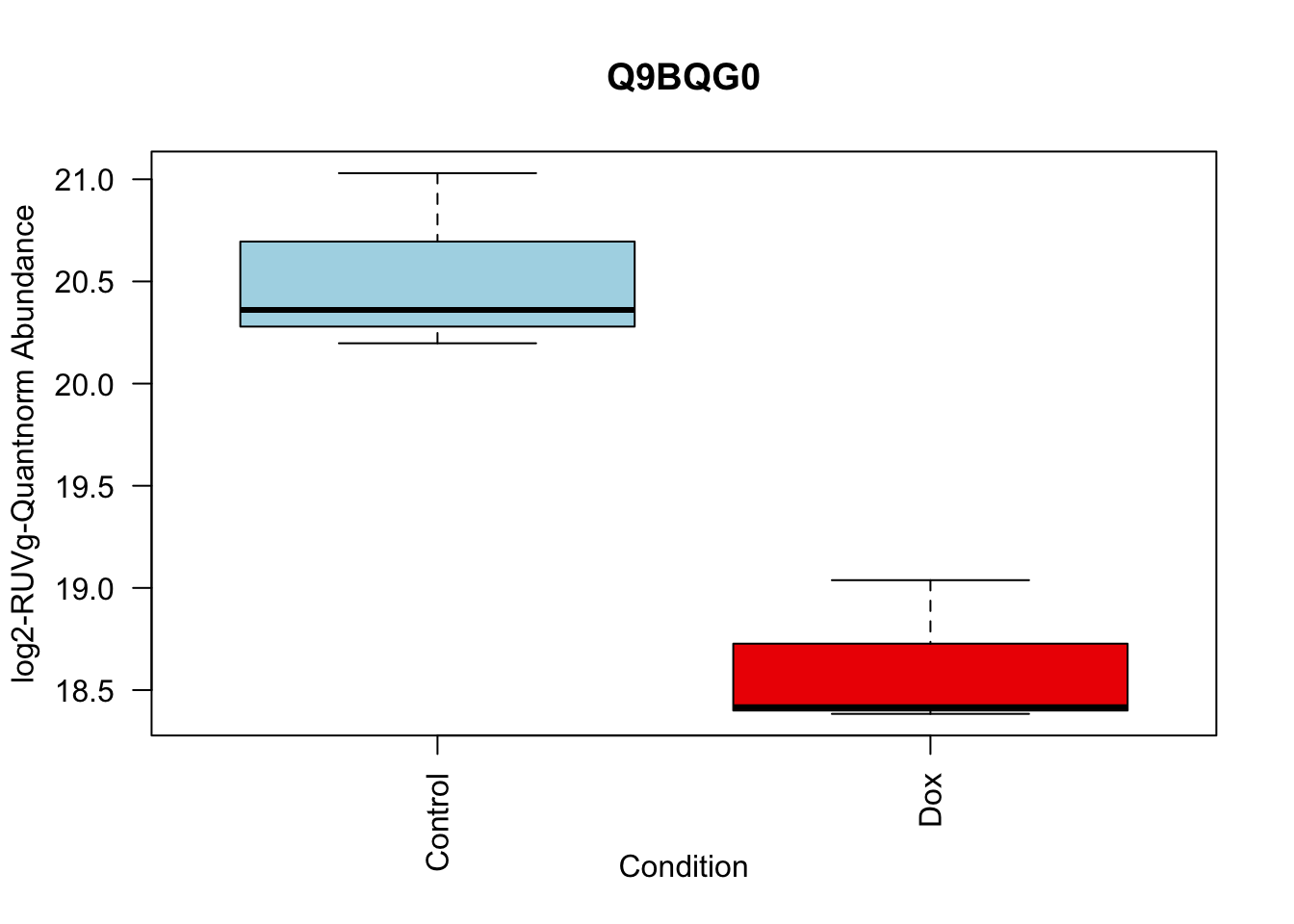
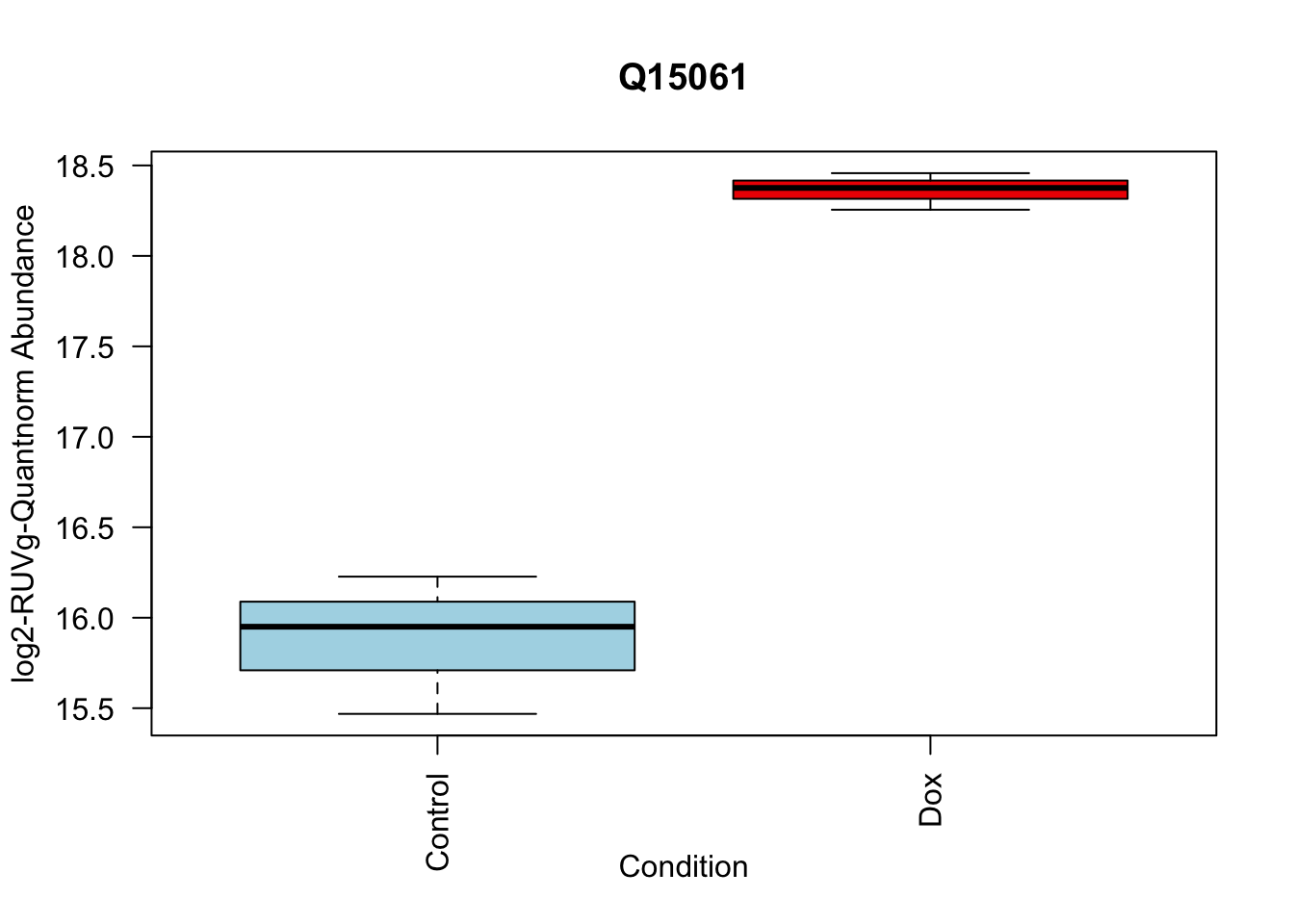
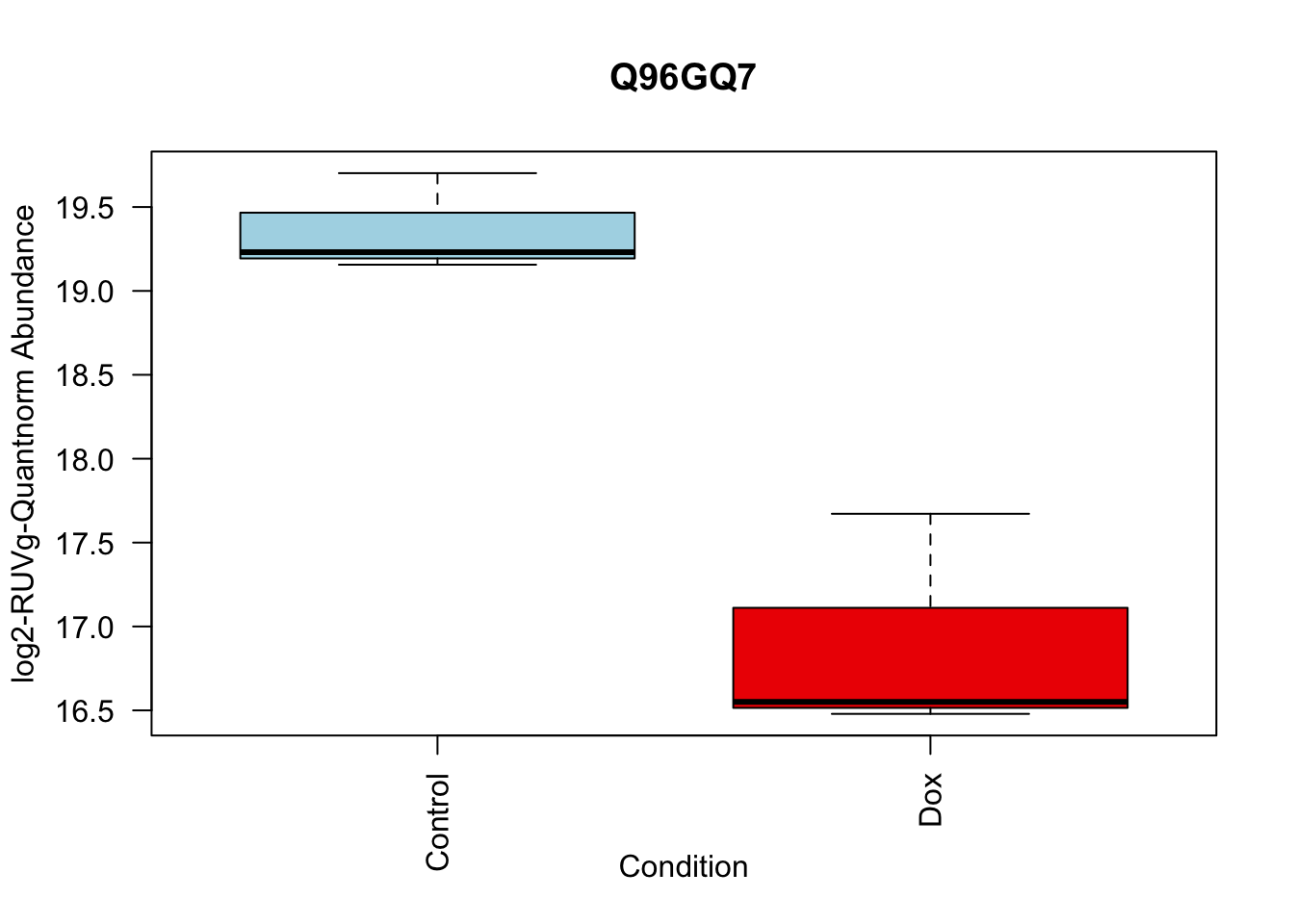
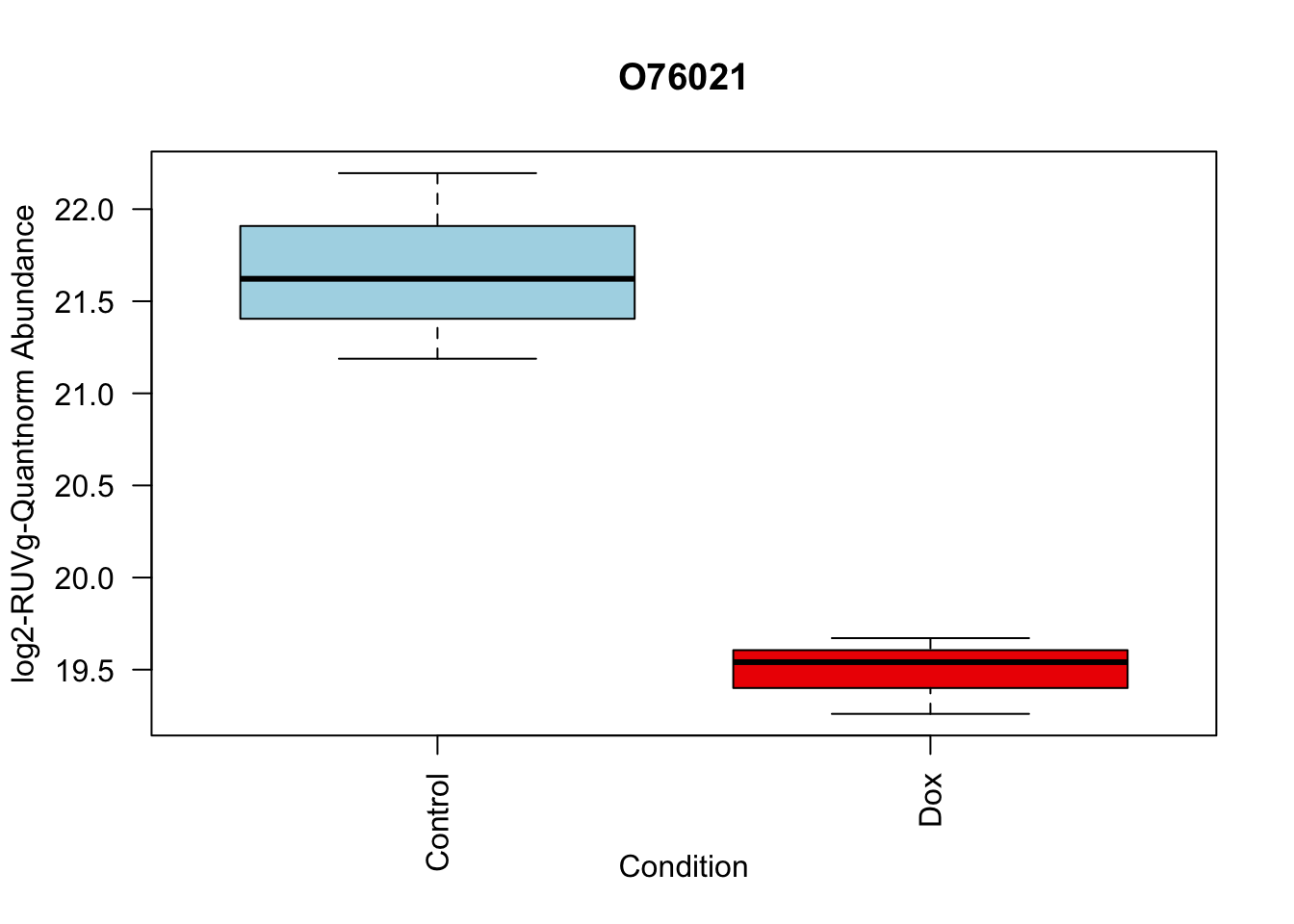
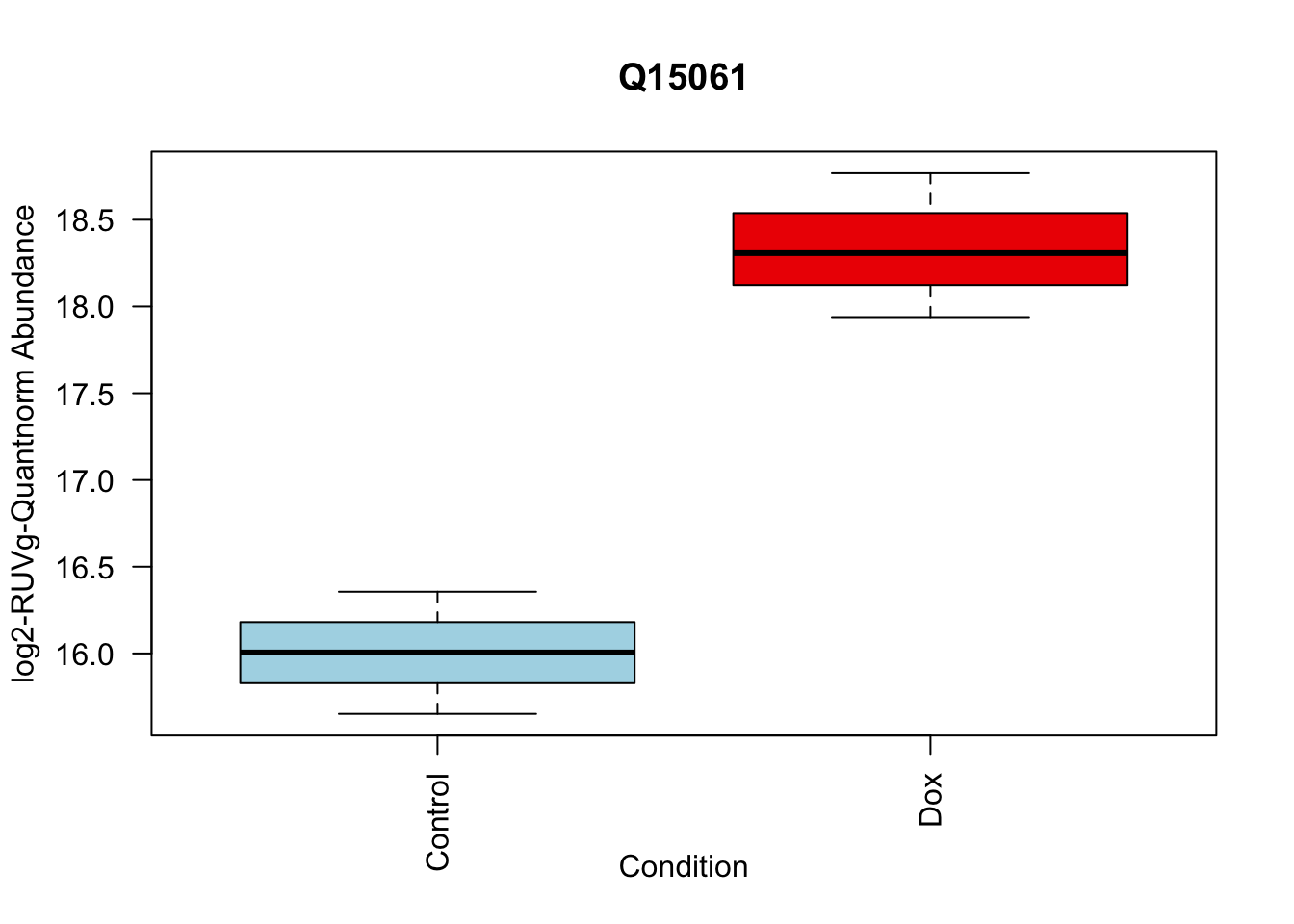
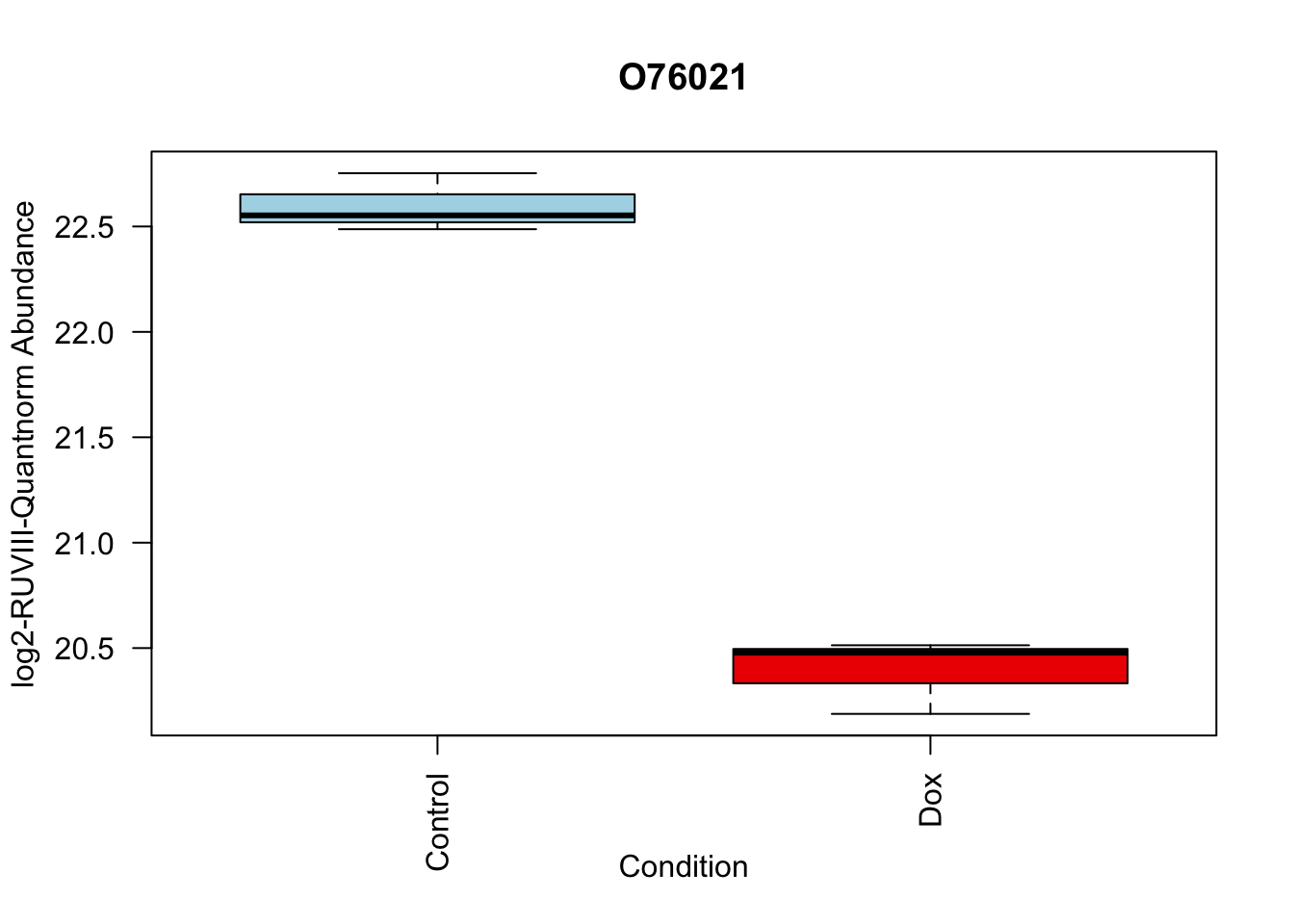
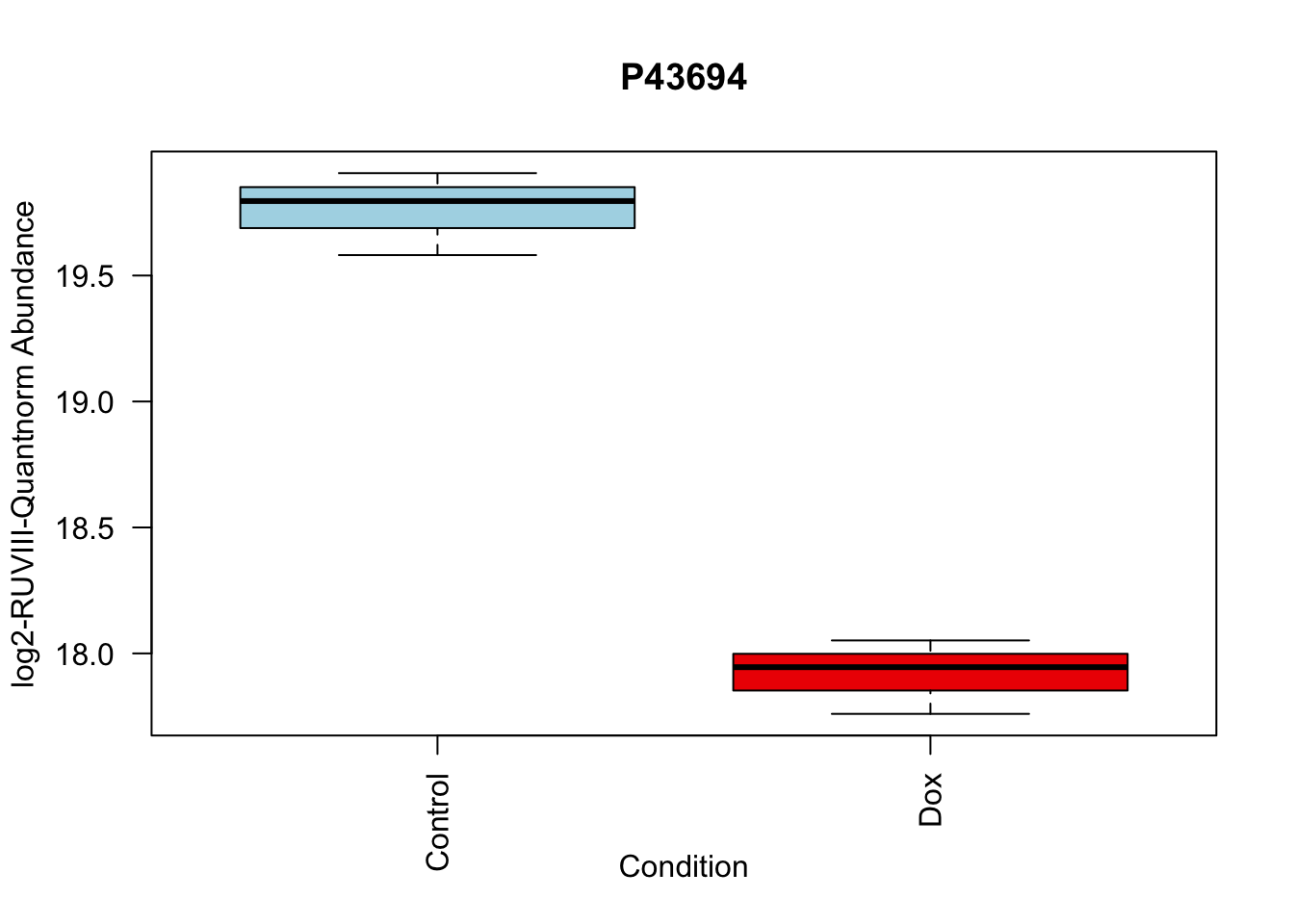
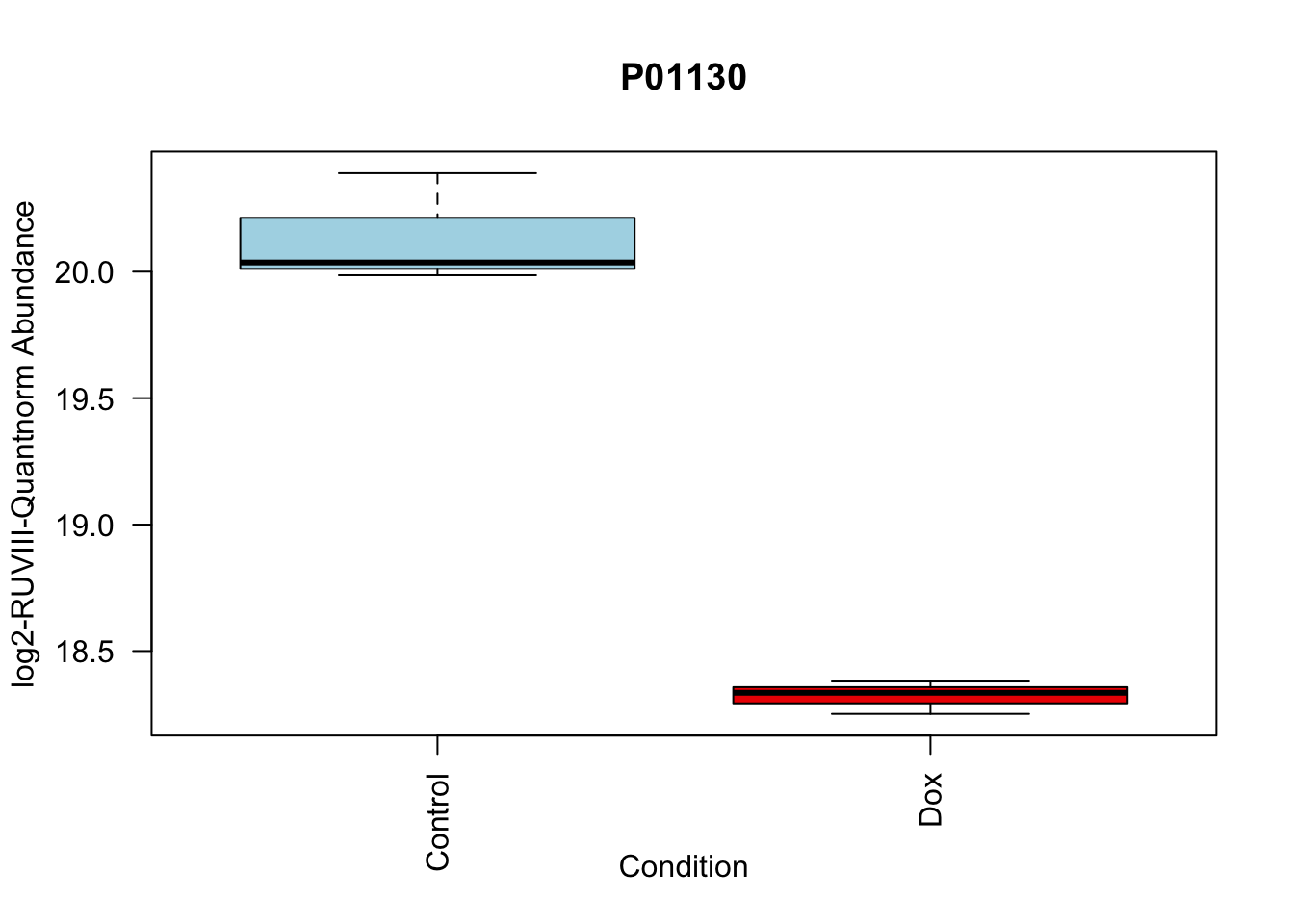
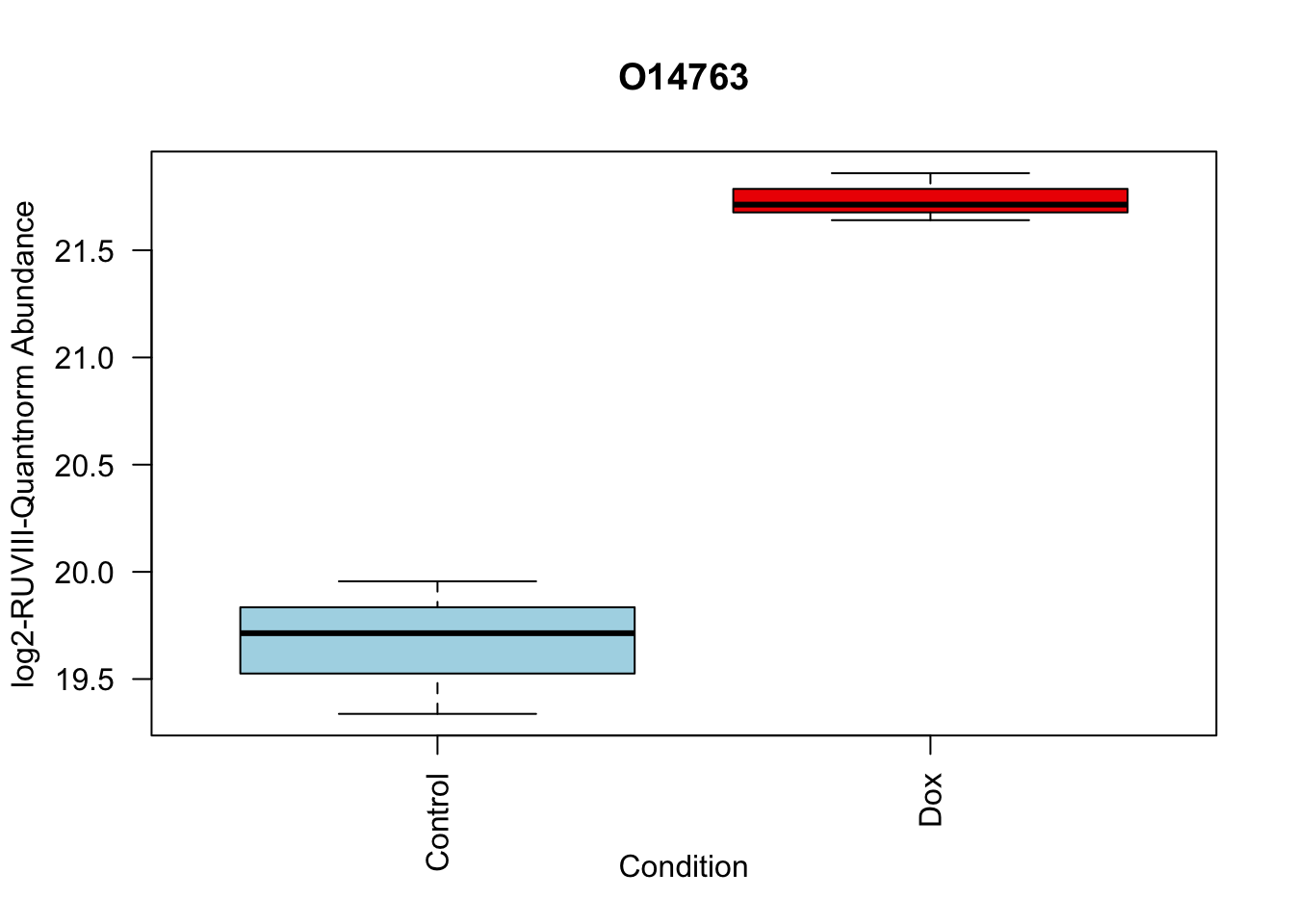
toptable_summary_DoxvNorm %>% head() logFC AveExpr t P.Value adj.P.Val B Protein
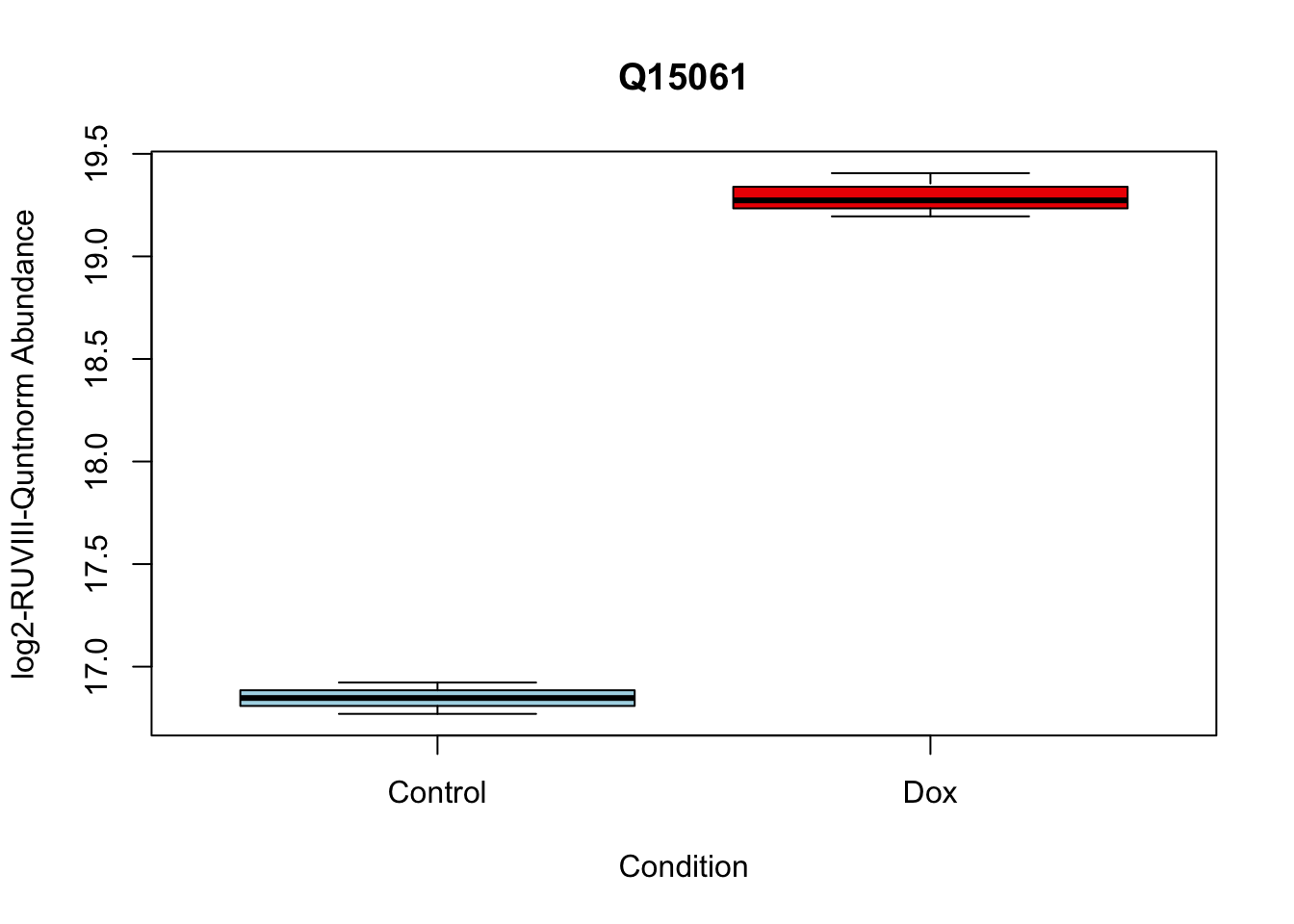
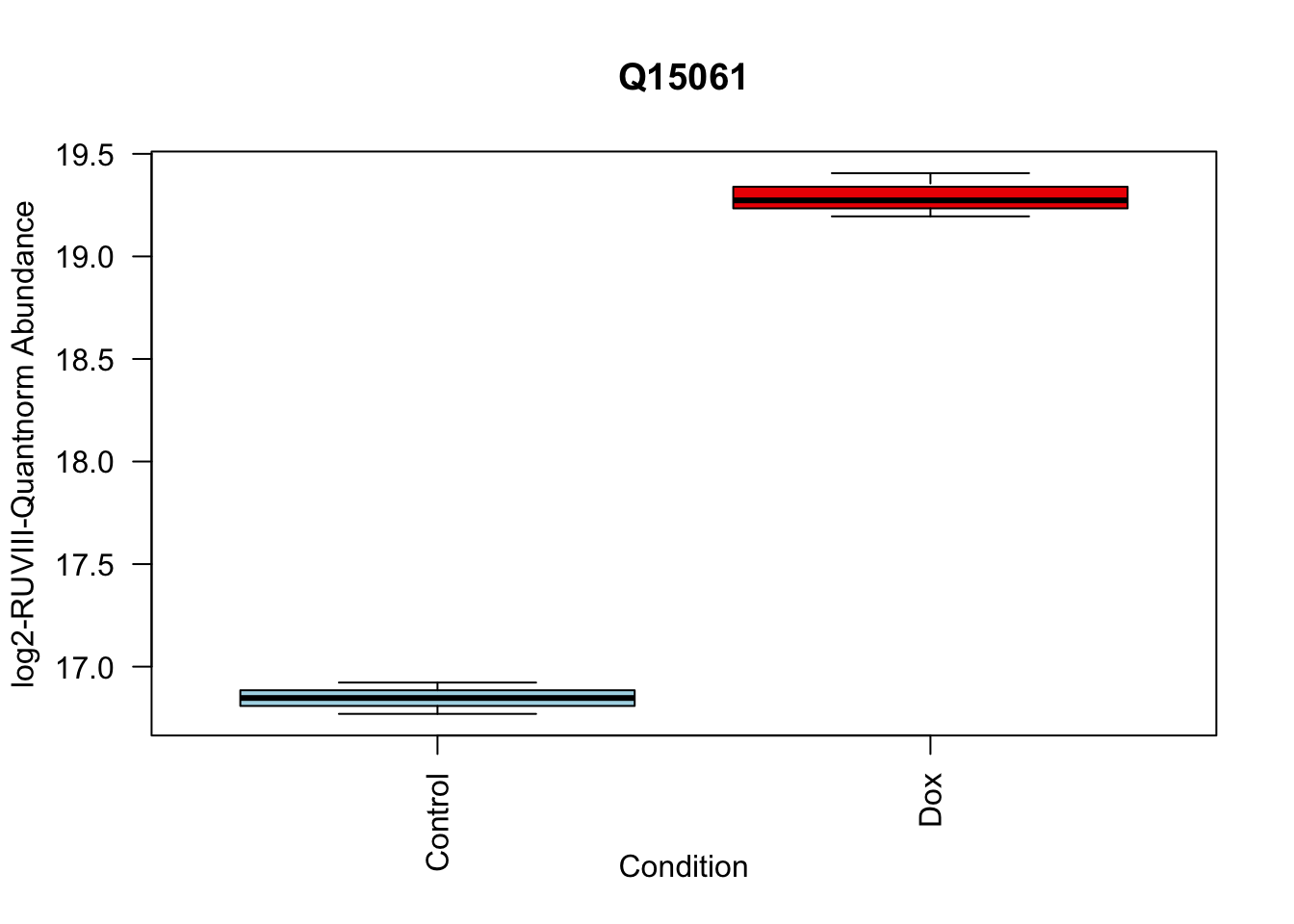
Q15061 2.444967 18.06896 22.98679 2.938337e-07 2.938337e-07 6.950800 Q15061
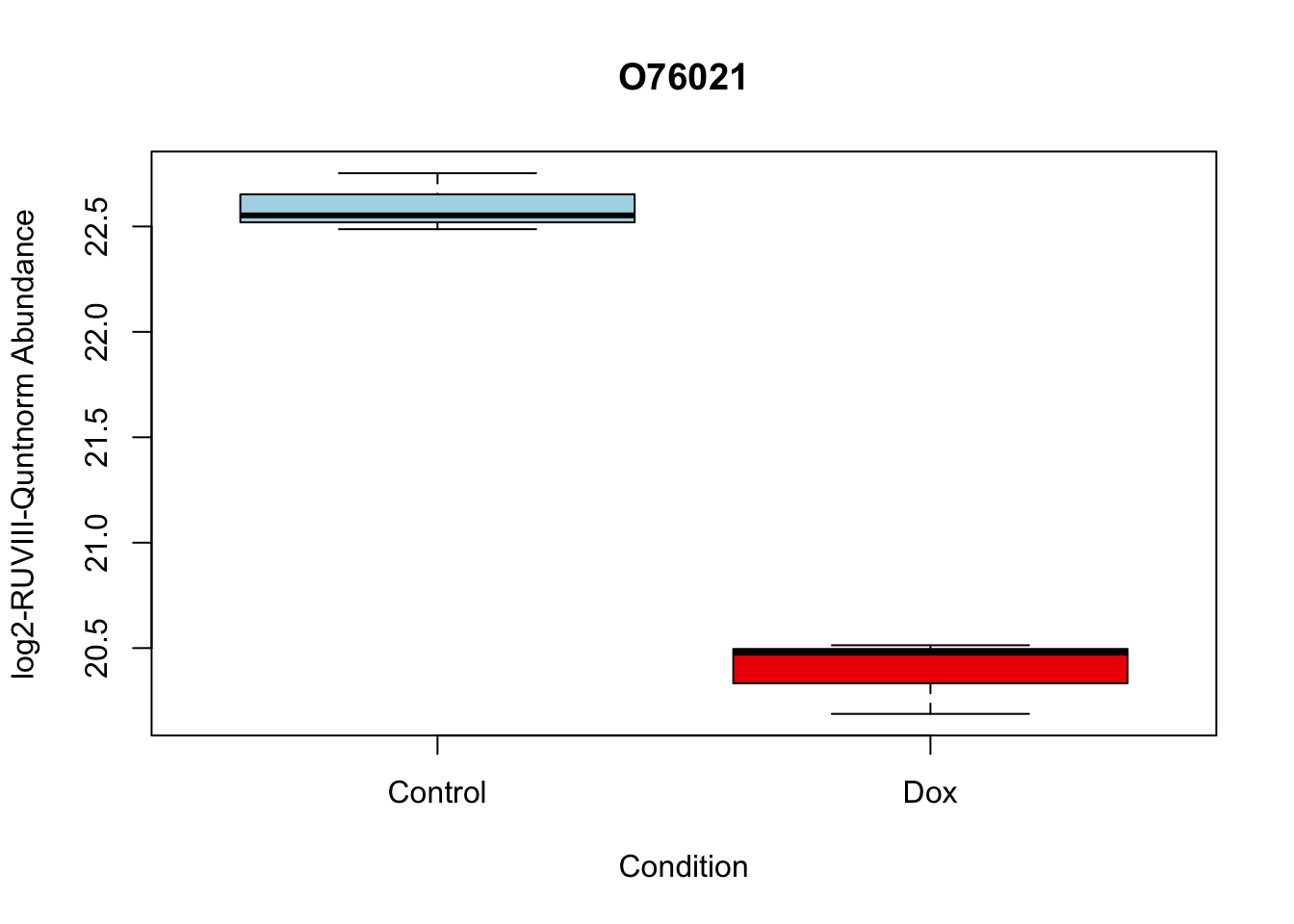
O76021 -2.204465 21.49534 -18.66029 1.173790e-06 1.173790e-06 5.995308 O76021
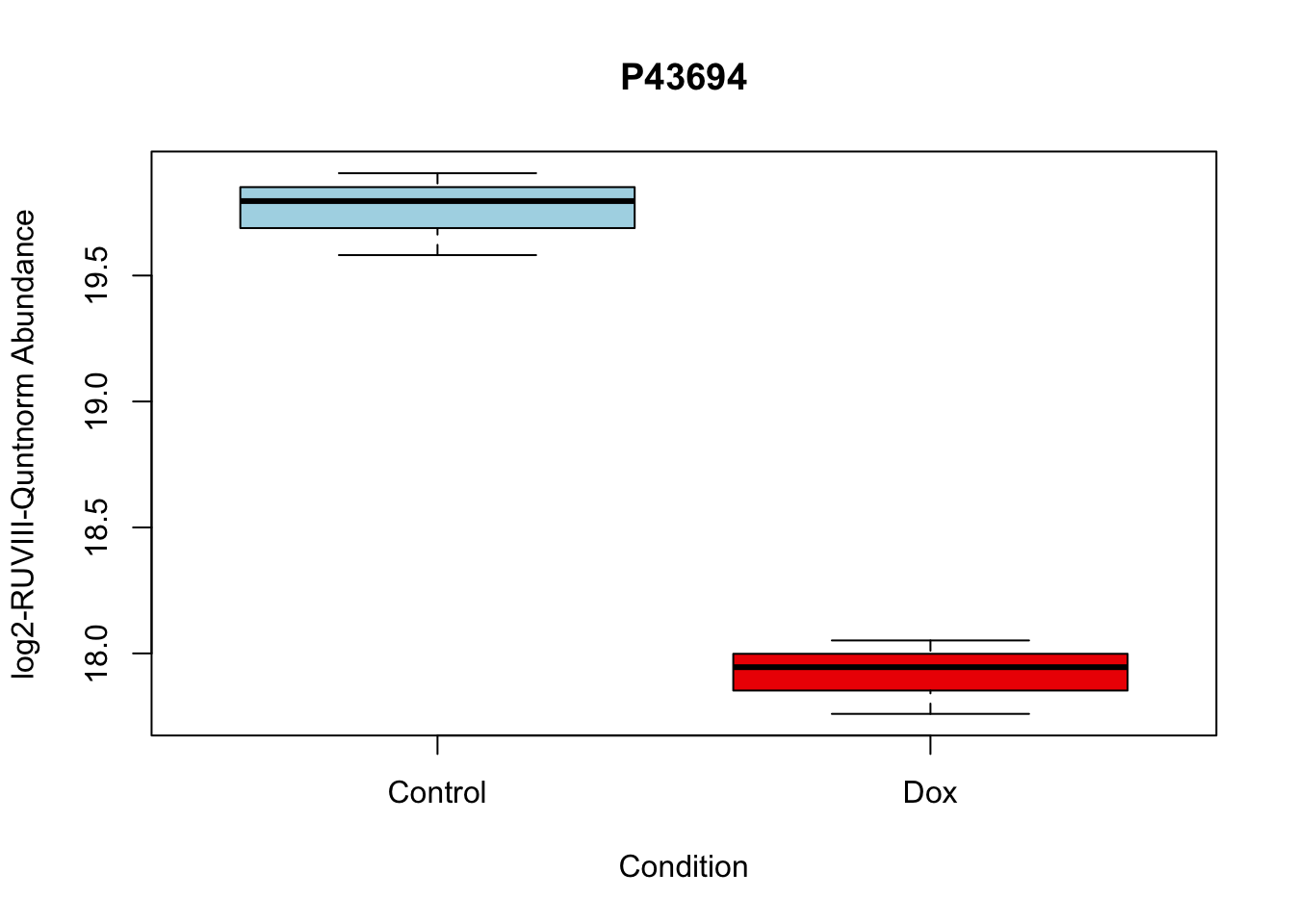
P43694 -1.842058 18.83998 -15.92359 2.895475e-06 2.895475e-06 5.308015 P43694
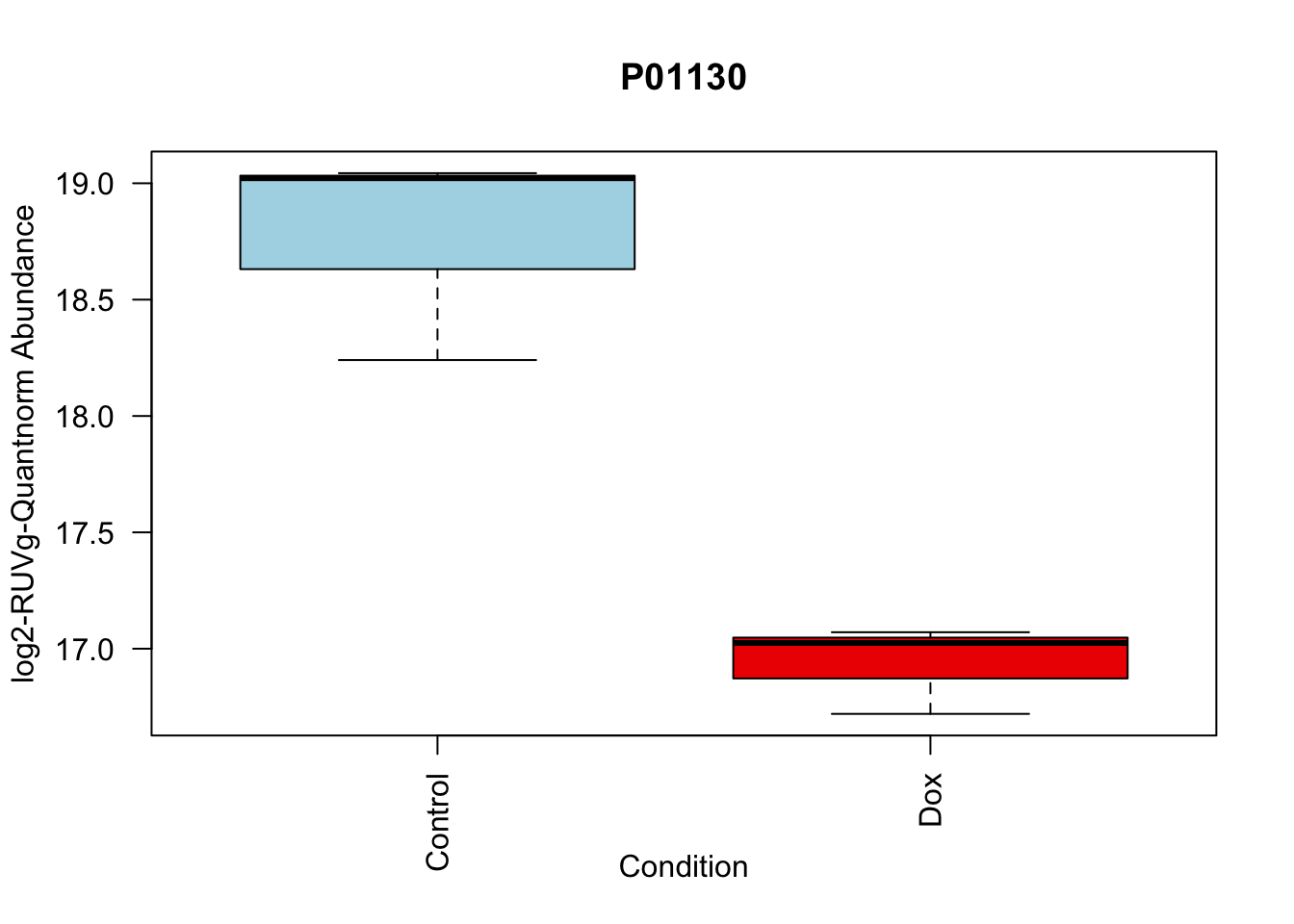
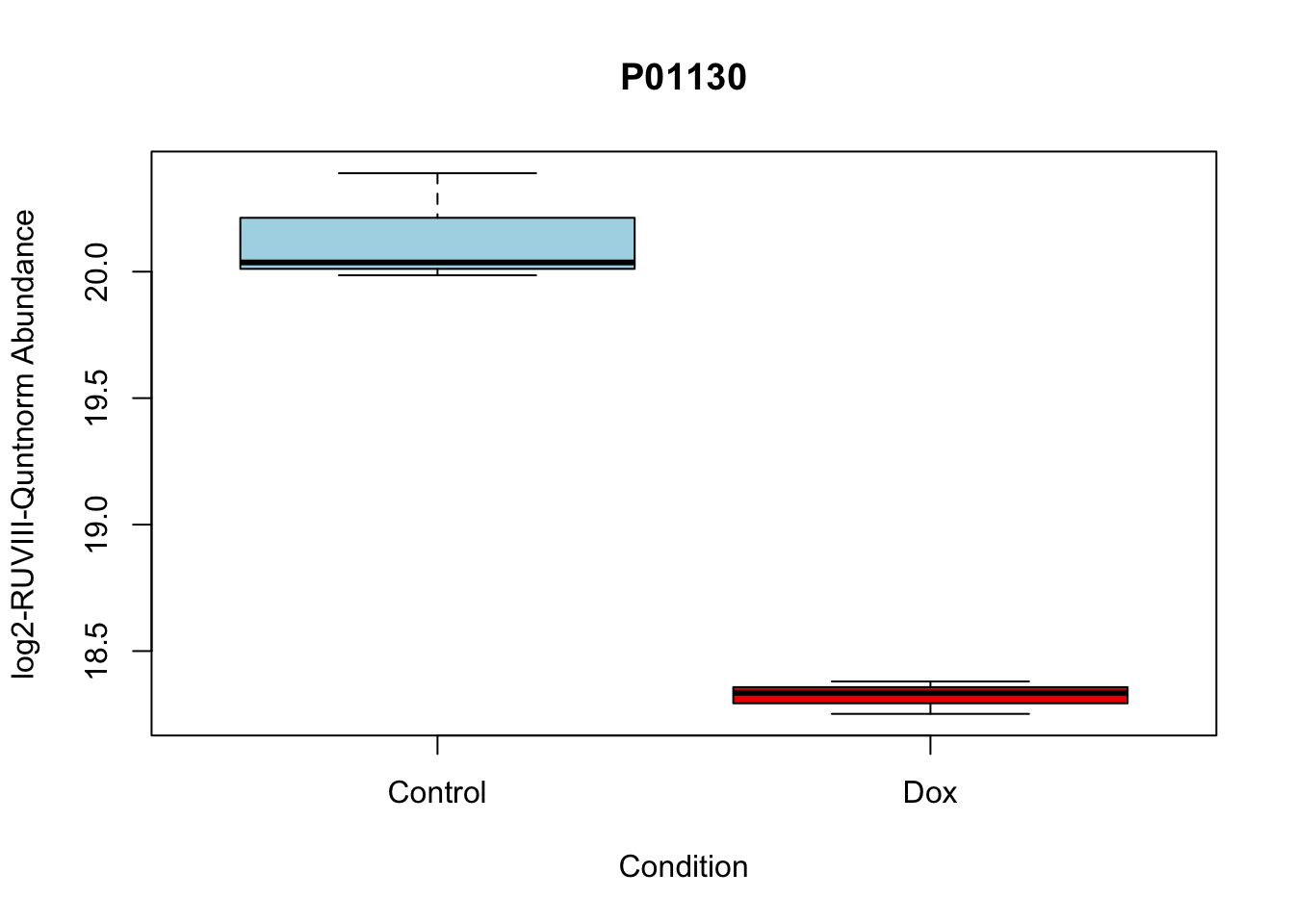
P01130 -1.815040 19.22969 -14.90808 4.560250e-06 4.560250e-06 4.929984 P01130
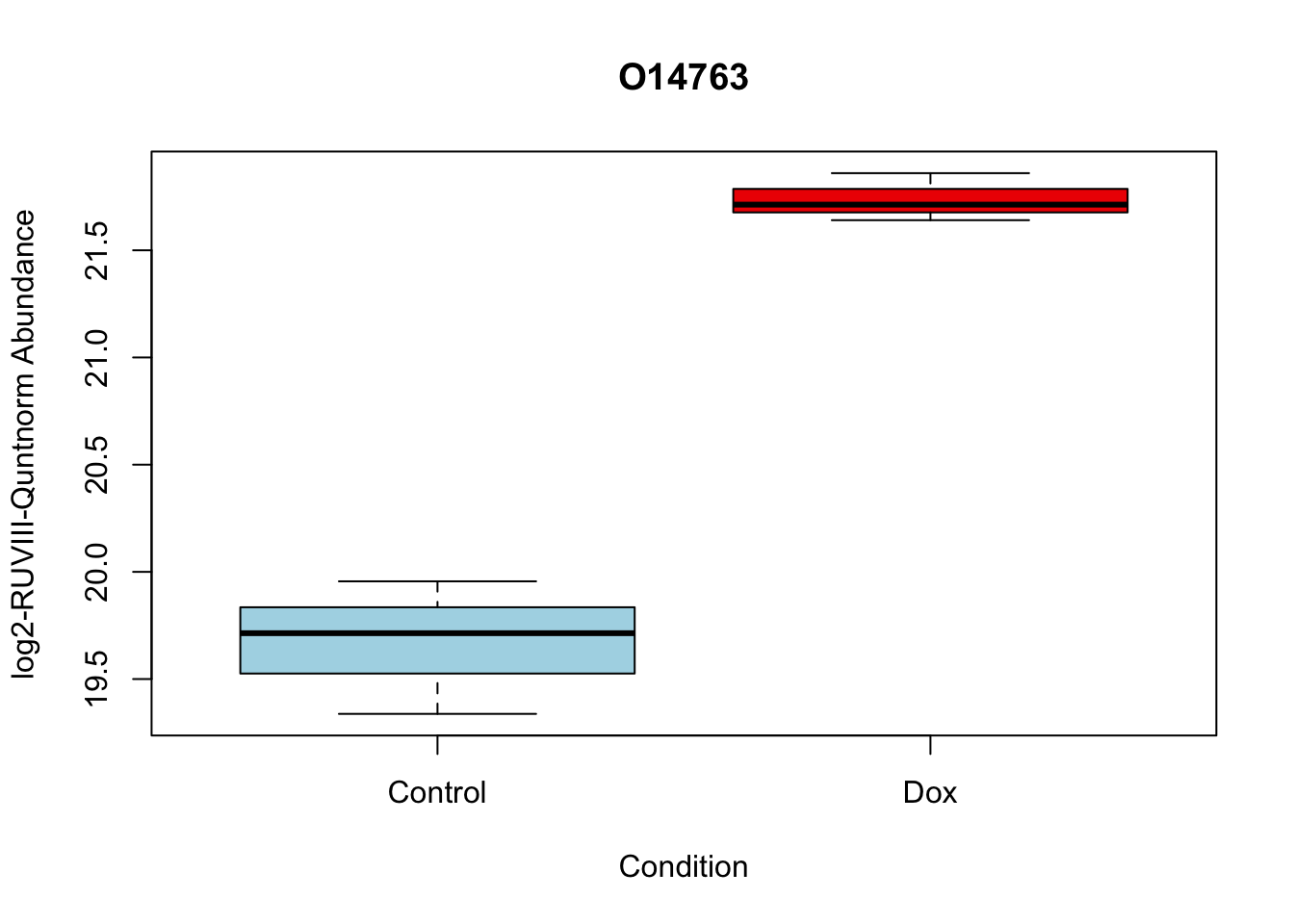
O14763 2.068618 20.70338 13.71850 7.513816e-06 7.513816e-06 4.502320 O14763
Q6EMK4 1.804211 18.96917 13.54255 8.117846e-06 8.117846e-06 4.434546 Q6EMK4# Volcano plots
# 1. Create a column to threshold P-values
toptable_summary_DoxvNorm <- toptable_summary_DoxvNorm %>% mutate(threshold_P = P.Value < 0.05)
# 2. Plot
ggplot(toptable_summary_DoxvNorm)+
geom_point(mapping = aes(x = logFC, y = -log10(P.Value), color = threshold_P))+
xlab("log2FC")+
ylab("-log10 nominal p-value")+
ylim(0, 7)+
xlim(-6, 6)+
theme(legend.position = "none",
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = rel(1.25)))+
theme_bw()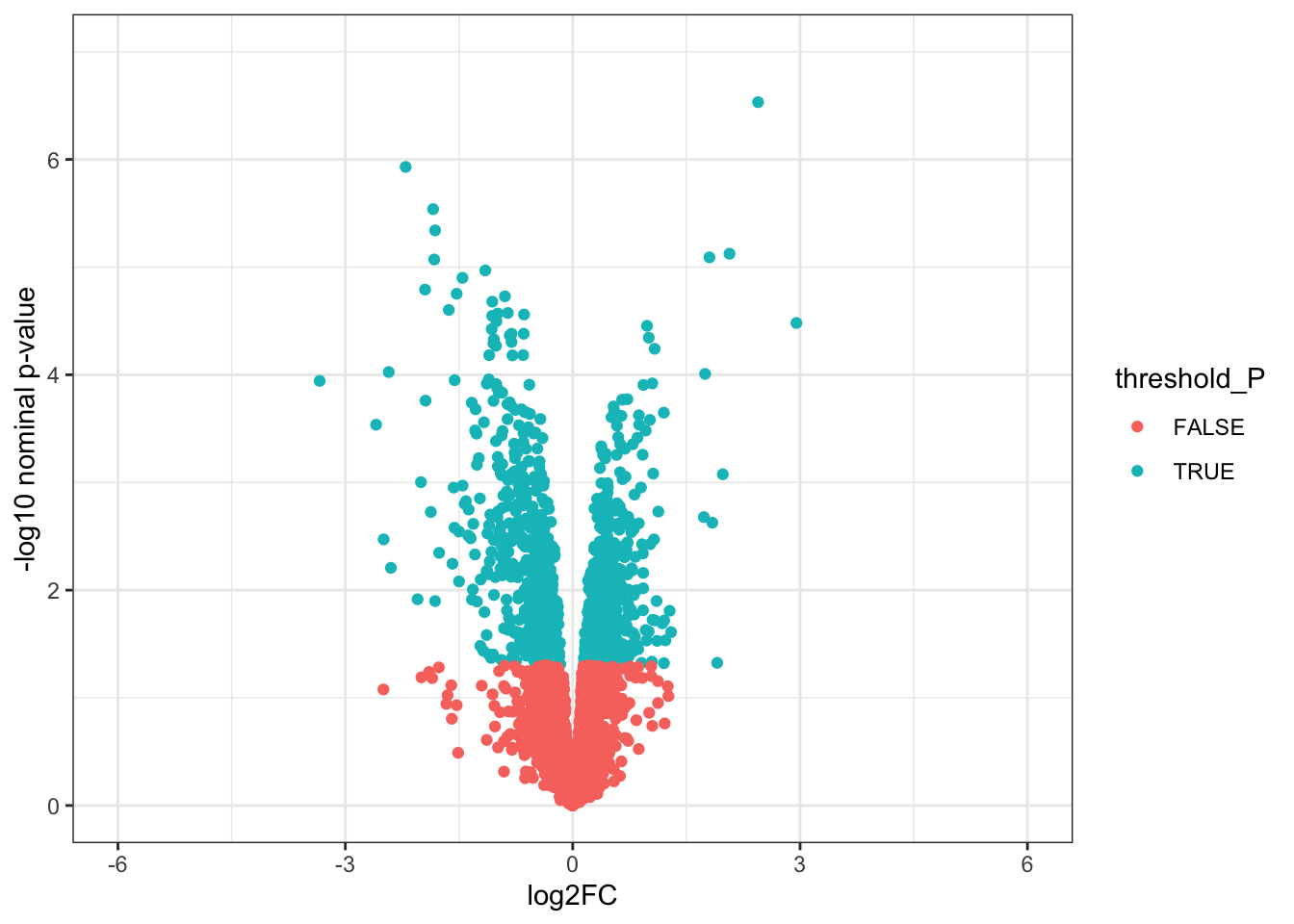
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Boxplots
toptable_summary_DoxvNorm[1:5,] %>% rownames()[1] "Q15061" "O76021" "P43694" "P01130" "O14763"# 1.
boxplot(Model_counts["Q15061",]~Meta_sub$Cond,
main = "Q15061",
xlab = "Condition",
ylab = "log2-RUVIII-Quntnorm Abundance",
col = c( "lightblue","red2")
)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# 2.
boxplot(Model_counts["O76021",]~Meta_sub$Cond,
main = "O76021",
xlab = "Condition",
ylab = "log2-RUVIII-Quntnorm Abundance",
col = c( "lightblue","red2")
)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# 3.
boxplot(Model_counts["P43694",]~Meta_sub$Cond,
main = "P43694",
xlab = "Condition",
ylab = "log2-RUVIII-Quntnorm Abundance",
col = c( "lightblue","red2")
)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# 4.
boxplot(Model_counts["P01130",]~Meta_sub$Cond,
main = "P01130",
xlab = "Condition",
ylab = "log2-RUVIII-Quntnorm Abundance",
col = c( "lightblue","red2")
)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# 5.
boxplot(Model_counts["O14763",]~Meta_sub$Cond,
main = "O14763",
xlab = "Condition",
ylab = "log2-RUVIII-Quntnorm Abundance",
col = c( "lightblue","red2")
)
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
# Make top_proteins your toptable summary
top_proteins <- toptable_summary_DoxvNorm[1:5, ] %>% rownames()
# Loop through each of the top proteins within the Model_counts
for (i in top_proteins) {
# Create a boxplot for each protein
boxplot(Model_counts[i, ] ~ Meta_sub$Cond,
main = i,
xlab = "Condition",
ylab = "log2-RUVIII-Quantnorm Abundance",
col = c("lightblue", "red2"),
las = 2) # las = 2 makes the axis labels perpendicular to the axis
}
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
| Version | Author | Date |
|---|---|---|
| e63bffa | Omar-Johnson | 2024-07-23 |
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur/Monterey 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] ruv_0.9.7.1
[2] DESeq2_1.36.0
[3] impute_1.70.0
[4] WGCNA_1.72-1
[5] fastcluster_1.2.3
[6] dynamicTreeCut_1.63-1
[7] BioNERO_1.4.2
[8] reshape2_1.4.4
[9] ggridges_0.5.4
[10] biomaRt_2.52.0
[11] ggvenn_0.1.10
[12] UpSetR_1.4.0
[13] DOSE_3.22.1
[14] variancePartition_1.26.0
[15] clusterProfiler_4.4.4
[16] pheatmap_1.0.12
[17] qvalue_2.28.0
[18] Homo.sapiens_1.3.1
[19] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[20] org.Hs.eg.db_3.15.0
[21] GO.db_3.15.0
[22] OrganismDbi_1.38.1
[23] GenomicFeatures_1.48.4
[24] AnnotationDbi_1.58.0
[25] cluster_2.1.4
[26] ggfortify_0.4.16
[27] lubridate_1.9.2
[28] forcats_1.0.0
[29] stringr_1.5.0
[30] dplyr_1.1.2
[31] purrr_1.0.2
[32] readr_2.1.4
[33] tidyr_1.3.0
[34] tibble_3.2.1
[35] ggplot2_3.4.3
[36] tidyverse_2.0.0
[37] RColorBrewer_1.1-3
[38] RUVSeq_1.30.0
[39] edgeR_3.38.4
[40] limma_3.52.4
[41] EDASeq_2.30.0
[42] ShortRead_1.54.0
[43] GenomicAlignments_1.32.1
[44] SummarizedExperiment_1.26.1
[45] MatrixGenerics_1.8.1
[46] matrixStats_1.0.0
[47] Rsamtools_2.12.0
[48] GenomicRanges_1.48.0
[49] Biostrings_2.64.1
[50] GenomeInfoDb_1.32.4
[51] XVector_0.36.0
[52] IRanges_2.30.1
[53] S4Vectors_0.34.0
[54] BiocParallel_1.30.4
[55] Biobase_2.56.0
[56] BiocGenerics_0.42.0
[57] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] rappdirs_0.3.3 rtracklayer_1.56.1 minet_3.54.0
[4] R.methodsS3_1.8.2 coda_0.19-4 bit64_4.0.5
[7] knitr_1.43 aroma.light_3.26.0 DelayedArray_0.22.0
[10] R.utils_2.12.2 rpart_4.1.19 data.table_1.14.8
[13] hwriter_1.3.2.1 KEGGREST_1.36.3 RCurl_1.98-1.12
[16] doParallel_1.0.17 generics_0.1.3 preprocessCore_1.58.0
[19] callr_3.7.3 RhpcBLASctl_0.23-42 RSQLite_2.3.1
[22] shadowtext_0.1.2 bit_4.0.5 tzdb_0.4.0
[25] enrichplot_1.16.2 xml2_1.3.5 httpuv_1.6.11
[28] viridis_0.6.4 xfun_0.40 hms_1.1.3
[31] jquerylib_0.1.4 evaluate_0.21 promises_1.2.1
[34] fansi_1.0.4 restfulr_0.0.15 progress_1.2.2
[37] caTools_1.18.2 dbplyr_2.3.3 geneplotter_1.74.0
[40] htmlwidgets_1.6.2 igraph_1.5.1 DBI_1.1.3
[43] ggnewscale_0.4.9 backports_1.4.1 annotate_1.74.0
[46] aod_1.3.2 deldir_1.0-9 vctrs_0.6.3
[49] abind_1.4-5 cachem_1.0.8 withr_2.5.0
[52] ggforce_0.4.1 checkmate_2.2.0 treeio_1.20.2
[55] prettyunits_1.1.1 ape_5.7-1 lazyeval_0.2.2
[58] crayon_1.5.2 genefilter_1.78.0 labeling_0.4.2
[61] pkgconfig_2.0.3 tweenr_2.0.2 nlme_3.1-163
[64] nnet_7.3-19 rlang_1.1.1 lifecycle_1.0.3
[67] downloader_0.4 filelock_1.0.2 BiocFileCache_2.4.0
[70] rprojroot_2.0.3 polyclip_1.10-4 graph_1.74.0
[73] Matrix_1.5-4.1 aplot_0.2.0 NetRep_1.2.7
[76] boot_1.3-28.1 base64enc_0.1-3 GlobalOptions_0.1.2
[79] whisker_0.4.1 processx_3.8.2 png_0.1-8
[82] viridisLite_0.4.2 rjson_0.2.21 bitops_1.0-7
[85] getPass_0.2-2 R.oo_1.25.0 ggnetwork_0.5.12
[88] KernSmooth_2.23-22 blob_1.2.4 shape_1.4.6
[91] jpeg_0.1-10 gridGraphics_0.5-1 scales_1.2.1
[94] memoise_2.0.1 magrittr_2.0.3 plyr_1.8.8
[97] gplots_3.1.3 zlibbioc_1.42.0 compiler_4.2.0
[100] scatterpie_0.2.1 BiocIO_1.6.0 clue_0.3-64
[103] intergraph_2.0-3 lme4_1.1-34 cli_3.6.1
[106] patchwork_1.1.3 ps_1.7.5 htmlTable_2.4.1
[109] Formula_1.2-5 mgcv_1.9-0 MASS_7.3-60
[112] tidyselect_1.2.0 stringi_1.7.12 highr_0.10
[115] yaml_2.3.7 GOSemSim_2.22.0 locfit_1.5-9.8
[118] latticeExtra_0.6-30 ggrepel_0.9.3 sass_0.4.7
[121] fastmatch_1.1-4 tools_4.2.0 timechange_0.2.0
[124] parallel_4.2.0 circlize_0.4.15 rstudioapi_0.15.0
[127] foreign_0.8-84 foreach_1.5.2 git2r_0.32.0
[130] gridExtra_2.3 farver_2.1.1 ggraph_2.1.0
[133] digest_0.6.33 BiocManager_1.30.22 networkD3_0.4
[136] Rcpp_1.0.11 broom_1.0.5 later_1.3.1
[139] httr_1.4.7 ComplexHeatmap_2.12.1 GENIE3_1.18.0
[142] Rdpack_2.5 colorspace_2.1-0 XML_3.99-0.14
[145] fs_1.6.3 splines_4.2.0 statmod_1.5.0
[148] yulab.utils_0.0.8 RBGL_1.72.0 tidytree_0.4.5
[151] graphlayouts_1.0.0 ggplotify_0.1.2 xtable_1.8-4
[154] jsonlite_1.8.7 nloptr_2.0.3 ggtree_3.4.4
[157] tidygraph_1.2.3 ggfun_0.1.2 R6_2.5.1
[160] Hmisc_5.1-0 pillar_1.9.0 htmltools_0.5.6
[163] glue_1.6.2 fastmap_1.1.1 minqa_1.2.5
[166] codetools_0.2-19 fgsea_1.22.0 utf8_1.2.3
[169] sva_3.44.0 lattice_0.21-8 bslib_0.5.1
[172] network_1.18.1 pbkrtest_0.5.2 curl_5.0.2
[175] gtools_3.9.4 interp_1.1-4 survival_3.5-7
[178] statnet.common_4.9.0 rmarkdown_2.24 munsell_0.5.0
[181] GetoptLong_1.0.5 DO.db_2.9 GenomeInfoDbData_1.2.8
[184] iterators_1.0.14 gtable_0.3.4 rbibutils_2.2.15